Abstract
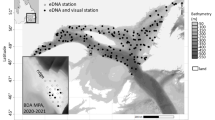
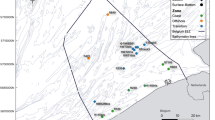
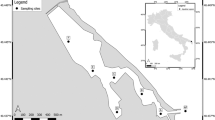
Rare species are difficult to observe in the wild, particularly in the ocean where large spatial scales and accessibility hinder effective sampling. Environmental DNA (eDNA) is a non-destructive, scalable sampling method with the potential to inform the distribution of rare species in marine ecosystems. We sample eDNA within the California Current ecosystem to estimate the distribution of eulachon (Thaleichthys pacificus), a threatened anadromous smelt ranging along the coastal Northeast Pacific. We amplify eulachon DNA from thousands of water samples collected at night across two years and more than 200,000 square kilometers along the U.S. west coast. We then use spatiotemporal models to derive quantitative estimates of eulachon DNA across space, depth, and time relative to environmental covariates. We find that eulachon DNA has a distribution weighted towards the ocean surface, spatially associated with major river mouths and productive offshore banks. Temperature and prey density are key covariates, with eulachon more likely to be found in warmer waters with higher prey concentrations. We discuss how our results can augment the information currently used in eulachon recovery planning, and describe the wide applicability of our statistical models for estimating distribution and abundance for other species of conservation concern.
Similar content being viewed by others
Data availability
All data and code required to reproduce our findings are archived and publicly available on Zenodo (https://doi.org/10.5281/zenodo.14721413). The source data behind the figures in the paper are available in the Zenodo archive. No genetic sequence data were generated in this study. All genetic analyses were performed via quantitative PCR (qPCR) using established, previously published primer sequences in Ramón-Laca et al.15 (also in Table 1) to quantify the expression of known targets, resulting in numerical data rather than producing sequence reads. Any remaining information can be obtained from the corresponding author upon reasonable request.
Code availability
The R code required to reproduce our findings are archived with the study data on Zenodo (https://doi.org/10.5281/zenodo.14721413).
References
Lomba, A. et al. Overcoming the rare species modelling paradox: a novel hierarchical framework applied to an Iberian endemic plant. Biol. Conserv. 143, 2647–2657 (2010).
Jeliazkov, A. et al. Sampling and modelling rare species: conceptual guidelines for the neglected majority. Glob. Change Biol. 28, 3754–3777 (2022).
Green, R. H. & Young, R. C. Sampling to detect rare species. Ecol. Appl. 3, 351–356 (1993).
Duarte, S., Simões, L. & Costa, F. O. Current status and topical issues on the use of eDNA-based targeted detection of rare animal species. Sci. Total Environ. 904, 166675 (2023).
Lafferty, K. Metabarcoding is (usually) more cost effective than seining or qPCR for detecting tidewater gobies and other estuarine fishes. PeerJ 12, e16847 (2024).
Shelton, A. O. et al. Environmental DNA provides quantitative estimates of Pacific hake abundance and distribution in the open ocean. Proc. R. Soc. B. 289, 20212613 (2022).
Jarman, S. N., Berry, O. & Bunce, M. The value of environmental DNA biobanking for long-term biomonitoring. Nat. Ecol. Evol. 2, 1192–1193 (2018).
Zizka, V. M. A., Koschorreck, J., Khan, C. C. & Astrin, J. J. Long-term archival of environmental samples empowers biodiversity monitoring and ecological research. Environ. Sci. Eur. 34, 40 (2022).
Keller, A. G., Grason, E. W., McDonald, P. S., Ramón-Laca, A. & Kelly, R. P. Tracking an invasion front with environmental DNA. Ecol. Appl. 32, e2561 (2022).
Sildever, S. et al. Eight years of weekly eDNA monitoring in the North-Western Pacific. Environ. DNA 5, 1202–1215 (2023).
Endangered and Threatened Wildlife and Plants Threatened status for southern distinct population segment of Eulachon. Fed. Reg. 50 CFR Part 223, 13012–13024 (2010).
Gustafson, R., Ford, M., Teel, D. & Drake, J. Status Review of Eulachon (Thaleichthys pacificus) in Washington, Oregon, and California. NOAA Technical Memorandum NMFS-NWFSC-105 (2010).
Gustafson, R. G. et al. Conservation status of eulachon in the California Current. Fish Fish. 13, 121–138 (2012).
Ward, E. J. et al. Using spatiotemporal species distribution models to identify temporally evolving hotspots of species co-occurrence. Ecol. Appl. 25, 2198–2209 (2015).
Ramón-Laca, A., Wells, A. & Park, L. A workflow for the relative quantification of multiple fish species from oceanic water samples using environmental DNA (eDNA) to support large-scale fishery surveys. PLOS ONE 16, e0257773 (2021).
Lellouche, J.-M. et al. Recent updates to the Copernicus Marine Service global ocean monitoring and forecasting real-time 1∕12° high-resolution system. Ocean Sci. 14, 1093–1126 (2018).
Gomez, F. A. et al. RC4USCoast: a river chemistry dataset for regional ocean model applications in the US East Coast, Gulf of Mexico, and US West Coast. Earth Syst. Sci. Data 15, 2223–2234 (2023).
Phillips, E. M. et al. Spatiotemporal variability of euphausiids in the California Current Ecosystem: insights from a recently developed time series. ICES J. Mar. Sci. 79, 1312–1326 (2022).
Yang, M.-S., Dodd, K., Hibpshman, R., & Whitehouse, A. Food Habits of Groundfishes in the Gulf of Alaska in 1999 and 2001. U.S. Dep. Commer., NOAA Tech. Memo. NMFS-AFSC-164 (2006).
Barraclough, W. E. Contribution to the marine life history of the Eulachon Thaleichthys pacificus. J. Fish. Res. Bd. Can. 21, 1333–1337 (1964).
Hay, D. & McCarter, P. B. Status of the eulachon Thaleichthys pacificus in Canada. Canadian Science Advisory Secretariat Research Document 2000/145, (2000).
Gustafson, R., Richerson, K. E., Somers, K. A., Tuttle, V. J. & McVeigh, J. T. Observed and Estimated Bycatch of Eulachon in the 2002-21 U.S. West Coast Fisheries. https://doi.org/10.25923/3611-EY18 (2023).
Hickey, B., Geier, S., Kachel, N. & MacFadyen, A. A bi-directional river plume: The Columbia in summer. Continental Shelf Research 25, 1631–1656 (2005).
Hickey, B. & Banas, N. Why is the Northern End of the California Current System So Productive? Oceanography 21, 90–107 (2008).
Barth, J. A., Pierce, S. D. & Castelao, R. M. Time-dependent, wind-driven flow over a shallow midshelf submarine bank. J. Geophys. Res. 110, C10S05 (2005).
Ressler, P. H. et al. The spatial distribution of euphausiid aggregations in the Northern California current during August 2000. Deep Sea Res. Part II52, 89–108 (2005).
Calambokidis, J. et al. 4. Biologically important areas for selected cetaceans within U.S. Waters – West Coast Region. Aquat. Mamm. 41, 39–53 (2015).
Derville, S., Barlow, D. R., Hayslip, C. & Torres, L. G. Seasonal, annual, and decadal distribution of three rorqual whale species relative to dynamic ocean conditions Off Oregon, USA. Front. Mar. Sci. 9, 868566 (2022).
Anderson, C. N. K. et al. Why fishing magnifies fluctuations in fish abundance. Nature 452, 835–839 (2008).
Emmett, R. L. Ecology of marine predatory and prey fishes off the Columbia River, 1998 and 1999. NOAA Technical Memorandum NMFS-NWFSC-51.
Litz, M. N. C., Emmett, R. L., Bentley, P. J., Claiborne, A. M. & Barceló, C. Biotic and abiotic factors influencing forage fish and pelagic nekton community in the Columbia River plume (USA) throughout the upwelling season 1999–2009. ICES J. Mar. Sci. 71, 5–18 (2014).
Bollens, S., Frost, B. & Lin, T. Recruitment, growth, and diel vertical migration of Euphausia pacifica in a temperate fjord. Mar. Biol. 114, 219–228 (1992).
Harrison, J. B., Sunday, J. M. & Rogers, S. M. Predicting the fate of eDNA in the environment and implications for studying biodiversity. Proc. R. Soc. B. 286, 20191409 (2019).
Hansen, B. K., Bekkevold, D., Clausen, L. W. & Nielsen, E. E. The sceptical optimist: challenges and perspectives for the application of environmental DNA in marine fisheries. Fish Fish. 19, 751–768 (2018).
Andruszkiewicz, E. A. et al. Modeling environmental DNA transport in the coastal ocean using Lagrangian particle tracking. Front. Mar. Sci. 6, 477 (2019).
Sutherland, B. J. G. et al. Population structure of eulachon (Thaleichthys pacificus) from Northern California to Alaska using single nucleotide polymorphisms from direct amplicon sequencing. Can. J. Fish. Aquat. Sci. 78, 78–89 (2021).
Hirayama, I. T., Wu, L. & Minamoto, T. Stability of environmental DNA methylation and its utility in tracing spawning in fish. Mol. Ecol. Resour. 24, e14011 (2024).
Piferrer, F. & Anastasiadi, D. Age estimation in fishes using epigenetic clocks: Applications to fisheries management and conservation biology. Front. Mar. Sci. 10, 1062151 (2023).
Lindgren, F., Rue, H. & Lindström, J. An explicit link between gaussian fields and gaussian markov random fields: the stochastic partial differential equation approach. J. R. Stat. Soc. Ser. B 73, 423–498 (2011).
Lindgren, F. et al. inlabru: software for fitting latent Gaussian models with non-linear predictors. Preprint at https://doi.org/10.48550/arXiv.2407.00791 (2024).
Anderson, S. C., Ward, E. J., English, P. A. & Barnett, L. A. K. sdmTMB: an R package for fast, flexible, and user-friendly generalized linear mixed effects models with spatial and spatiotemporal random fields. Preprint at https://doi.org/10.1101/2022.03.24.485545 (2022).
Kristensen, K., Nielsen, A., Berg, C. W., Skaug, H. & Bell, B. M. TMB: Automatic Diff erentiation and Laplace Approximation. J. Stat. Softw. 70, 1–21 (2016).
Acknowledgements
The authors gratefully acknowledge the survey expertise and support of NOAA’s Pacific Hake survey team and the NOAA Ship Bell M. Shimada personnel for supporting the collection of eDNA samples on the West Coast Hake Survey 2019-2021. In addition, the authors acknowledge L. Park and R. Gustafson for their leadership in the early stages of the development of this study; without their support this work would not have been possible. This work was supported in part by the NOAA Fisheries Omics Strategic Initiative (2019-2023).
Author information
Authors and Affiliations
Contributions
Conceptualization: O.R.L., A.O.S., K.M.N., and R.P.K. Methodology: O.R.L., A.O.S., R.P.K., and E.J.W. Data analysis: O.R.L. and E.J.W. Data curation: A.R.-L., A.O.S., K.M.N., R.P.K., E.M.P., and A.W. Writing and editing: O.R.L., A.O.S., A.R.-L., K.M.N., E.J.W., E.M.P., J.E.Z., A.W., and R.P.K.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Mads Reinholdt Jensen, Cristina Claver, and the other anonymous reviewers for their contribution to the peer review of this work. Primary handling editors: Linn Hoffmann, Aylin Bircan, and Rupali Sathe. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, O.R., Shelton, A.O., Ramón-Laca, A. et al. Mapping the marine distribution of eulachon (Thaleichthys pacificus) in the Northeast Pacific using environmental DNA. Commun Biol (2026). https://doi.org/10.1038/s42003-026-09733-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-026-09733-5