Abstract
While the abilities of language models are thoroughly evaluated in areas like general domains and biomedicine, academic chemistry remains less explored. Chemical QA tools also play a crucial role in both education and research by effectively translating complex chemical information into an understandable format. Addressing this gap, we introduce ScholarChemQA, a large-scale QA dataset constructed from chemical papers. Specifically, the questions are from paper titles with a question mark, and the multi-choice answers are reasoned out based on the corresponding abstracts. This dataset reflects typical real-world challenges, including an imbalanced data distribution and a substantial amount of unlabeled data that can be potentially useful. Correspondingly, we introduce a ChemMatch model, specifically designed to effectively answer chemical questions by fully leveraging our collected data. Experiments show that Large Language Models (LLMs) still have significant room for improvement in the field of chemistry. Moreover, ChemMatch significantly outperforms recent similar-scale baselines: https://github.com/iriscxy/chemmatch.
Similar content being viewed by others
Introduction
Question Answering (QA) models have emerged as crucial tools for acquiring knowledge and evaluating domain-specific abilities. For example, QA models are designed to provide precise answers to a wide range of queries, thus assisting in the dissemination of information and the enhancement of learning processes1,2,3,4,5. Correspondingly, to examine and evaluate the accuracy of the given answers6,7,8,9, propose different QA datasets to rank the abilities of various language models and find that these models have flaws in different ways. Question answering is often framed as a judgment task. For example, in the BoolQ task10, where a user poses questions about a document, the QA model responds with either “yes” or “no,” as illustrated in Fig. 1a. This type of judgment can be challenging even in general contexts. For instance10, found that answering natural questions is surprisingly difficult, as they frequently require a deep understanding of context, nuances, and specific details within the document. In scientific domains, judgment tasks are also a common format. In studies such as11,12, the task involves either supporting or refuting a scientific claim. QA tasks in scholarly fields are particularly demanding, as scientific papers often contain specialized terminology that can be challenging to understand even for researchers13,14,15. A number of domain-specific QA datasets are proposed in the biomedical domain16,17,18. For example19, proposes a multi-choice biomedical QA dataset collected from PubMed papers, and20 collects a multiple-choice dataset to classify which disease the patient has.21 proposes LiteratureQA, a QA corpus consisting of papers in the computer science domain with human-engineered questions.
Comparison of (a) general domain QA dataset BoolQ, (b) chemical domain dataset KGQA, and (c) our ScholarChemQA dataset. Our dataset is sourced from chemical research papers, in contrast to previous chemical datasets, which were artificially constructed. Our dataset contains text rich in domain-specific information, making it highly suitable for evaluation.
However, the domain of chemical QA has not been explored as extensively as other scientific fields, such as biology19,22. Recent interdisciplinary research efforts have increasingly employed language models as tools in chemistry23,24,25,26. In the meantime, chemical QA systems provide quick, accurate access to essential chemical information, aiding in the resolution of complex problems, understanding reactions, and the development of new materials, thus supporting innovation and informed decision-making in chemistry-related fields27,28,29,30,31. For example32, proposes multimodal multiple-choice questions on different science topics, along with annotations of their answers, corresponding lectures, and explanations. The dataset most closely related to our work is KGQA28, as shown in Fig. 1b. KGQA is based on a chemical knowledge graph and relies on a template-based approach to generate QA pairs. However, this approach lacks the diversity found in real-world language and tends to focus primarily on foundational chemical concepts rather than complex, practical research questions. Additionally, KGQA depends heavily on a human-constructed knowledge graph, limiting its adaptability and scope for broader research applications.
In contrast to previous work, this study introduces a chemical research QA benchmark to evaluate and improve the chemical QA capabilities of language models by leveraging the large-scale scholarly chemical papers that are readily available. Each year, there are over 500,000 new publications in the field of chemistry, as reported by the Web of Science, making it an excellent resource to start with. The QA pairs in these papers originate from research-investigated problems rather than being artificially created for evaluation, thus holding greater relevance and applicability to practical scenarios in the field of chemistry. Concretely, in this paper, we propose ScholarChemQA, a chemical QA dataset for answering research questions with multi-choice between yes, no, and maybe. Firstly, we collected over a million titles and abstracts related to chemistry from academic platforms. Through a rigorous selection process, we curated 40k QA pairs where each title, framed as a question, can be answered using the aforementioned options. Out of these, 1k pairs were hand-labeled for training, validation, and testing, with yes/no/maybe constituting 65.8%, 21.2%, and 13.0%, respectively. The ‘yes’ and ‘no’ labels indicate if the abstract’s experiments support or refute the conclusion, and the ‘maybe’ label serves as a nuanced indicator for ambiguous or mixed evidence situations. Besides, to enrich our dataset, we converted an additional 4k titles from statement format into yes/no questions. An example case from our dataset is shown in Fig. 1c. To correctly answer the question, the model should have a foundational understanding of the behavior of a two-dimensional hole gas, the principles of GaAs quantum wells, and the concept of phase separation. Semantic reasoning skills are also indispensable to interpret the ‘coexistence of two phases’ as the concurrent existence of the mentioned metallic and insulating phases. The benefits of our datasets are multi-faceted. Firstly, it is a chemical QA dataset for research purposes, encompassing a wide range of topics from basic concepts to complex chemical processes. Secondly, it requires complex reasoning and in-depth semantic analysis to deduce the answer. Thirdly, ScholarChemQA sets a new benchmark for AI in real-world, academic contexts, enhancing AI-driven exploration and discovery in chemistry.
For experiments, we first evaluate the performance of LLMs on ScholarChemQA. Results show that even the advanced GPT-3.5 model achieves only 54% accuracy, highlighting the difficulties faced by LLMs in understanding research papers filled with complex terminology. Recognizing the need for improvement and more accessible resources, we aim to use our collected chemical QA dataset to develop a smaller, more precise model. The first challenge here is that the dataset exhibits an imbalanced attribute, where just 13% of cases belong to the ‘maybe’ minority class. This is a commonly observed characteristic in real-world datasets, as noted in previous studies33. This imbalance becomes more pronounced when including the automatically annotated yes/no set. The second challenge involves the incorporation of a substantial amount of unlabeled data. Hence, in this paper, we introduce ChemMatch, a chemical question-answering model with label rebalance, pseudo label calibration, and dual augmentation to address the above challenges. Generally, our ChemMatch follows the semi-supervised paradigm, generating pseudo-labels for unlabeled data and training the model to predict these labels using augmented data. We first address the issue of imbalanced label distribution by re-weighting the instance-wise loss based on the inverse frequency of each class. The pseudo label calibration seeks to align pseudo-label estimates with a desired ground truth distribution. To alter unlabeled samples for creating diverse augmentations, we propose a SoftMix operation that generates both question- and context-side augmentation, not in the input space, but in their representation space. Our experimental results demonstrate that our proposed model significantly outperforms models of a similar scale and LLM, marking a step forward in domain-specific QA model development.
Our main contributions can be summarized as follows:
-
We collected ScholarChemQA, a chemical QA dataset for answering research questions. This benchmark can be used to evaluate the chemistry domain capabilities of AI models.
-
We assess recent LLMs including Llama2-70B, GPT-3.5, and GPT-4 on ScholarChemQA, revealing their limitations in comprehending chemical research papers and delivering precise answers.
-
We propose an open-source, and computationally efficient model ChemMatch. ChemMatch significantly outperforms the advanced GPT-3.5 and GPT-4 models, providing a valuable tool for acquiring chemical-related knowledge.
ScholarChemQA Dataset
In this section, we introduce our data collection process and some key attributes of our collected data.
Data collection
Data sources
To compile a comprehensive collection of chemical papers, we utilized multiple academic publishing sources including Elsevier and Springer. The overall process is illustrated in Fig. 2a. Firstly, by employing a combination of publisher APIs for databases such as Scopus, ScienceDirect, Springer Nature, Cross-Ref, and Lens34, we collected approximately 10 million abstracts and titles centered on chemistry-related studies from 2000 to 2023. Then, we specifically selected papers that have question marks in their titles to build the QA dataset. This is because we can automatically obtain natural scholarly questions, and the corresponding answer is usually found within the abstract or the main content of the paper. By focusing on papers with question marks in their titles, we aim to capture a diverse set of research questions that are directly relevant to the field of chemistry. This approach allows us to construct a dataset that is rich in domain-specific questions and answers, providing a valuable resource for training and evaluating question-answering models in the scientific domain. In this work, we employ a multi-choice setting, where the questions are answered with ‘yes’, ‘no’, or ‘maybe’. This approach simplifies the response format and allows for a more straightforward evaluation of the question-answering model’s performance. By restricting the answers to these three options, we can focus on the model’s ability to understand and categorize the information presented in the text, making it easier to assess its accuracy and reliability in a controlled setting.
a Illustration of data crawling process. b Topic distribution of ScholarChemQA. c Proportional relationships between corresponding question types and reasoning types. Different question types correspond to different reasoning types, showcasing the diversity of our dataset. 71.5% of the questions require chemical knowledge for answering, showing the difficulty of our chemical question-answering tasks.
Note that not all questions can be answered with a yes/no/maybe response. To handle this, we followed a rule-based approach, where we excluded questions that start with interrogative words (e.g., wh-words) or involve selecting from multiple entities. During our initial investigation, we found that approximately 10% of the abstracts contained a conclusion subsection that could be considered as the response to the associated question. To enhance the challenge of reasoning, we excluded this section from our context. Finally, we obtained 40k cases that cover various topics, and the distribution of papers from various sources is shown in Fig. 2b.
Expert annotation and quality control
Since the original dataset lacked answer labels for the question titles, we conducted an expert annotation process to collect a labeled dataset. The annotation criterion was as follows: We choose to annotate a question with ‘yes’ when the experiments and results of the paper substantiate it. Conversely, we use ‘no’ when they contradict or refute the statement. A ‘maybe’ is annotated in two scenarios: (1) when the paper outlines conditions in which the answer could be both true and false, or (2) when multiple interventions, observations, etc., are inquired about, and the answer holds true for some but not all of them. It is crucial to recognize that these answers are not universal truths, but rather depend on the specific context provided in the research paper. We employed four PhD annotators, each with a background in chemistry, to individually label 525 instances, yielding two annotations for each case and a labeled dataset consisting of 1050 instances. One annotator had access to the conclusion part, reducing the need for extensive reasoning, while the other annotator was not provided with the conclusion part, requiring deeper reasoning from the available context. This separation process ensured both annotation and dataset quality. When there was disagreement in the labeling, a third annotator facilitated discussions to achieve consensus among the two initial annotators. The initial labeling yielded a Kappa score of 0.62, indicating substantial agreement, and the final discussion phase ensured an overall high quality of the data. The statistics are shown in Table 1.
For the human-annotated cases, the train, validation, and test sets consist of 500, 50, and 500 samples, respectively. Next, we collect additional automatically annotated training cases.
Automatic annotation
To further enrich our dataset, we used a simple heuristic to collect noisily-labeled instances. We began by selecting papers with statement titles that followed specific Part-Of-Speech (POS) tagging structures (NP-(VBP/VBZ)) based on the Stanford POS tagging scheme35. We then transformed the statement titles into questions by employing a simple method, which involved inserting copulas like ‘is’ or auxiliary verbs such as ‘does’ at the beginning of the sentence. We also ensured that the transformed sentences were coherent, making necessary adjustments like adding question marks. The yes/no answer was then determined based on whether the verb (VB) in the sentence was negated. For example, the title ‘Current fossil fuel infrastructure does not yet commit us to 1.5 ∘C warming’ is changed to ‘Does the current fossil fuel infrastructure commit us to 1.5 ∘C warming?’ with answer ‘No’. In cases where the complex titles involve commas or colons, we relied on GPT-4 to automatically convert them into appropriate question formats. In a random sampling of 200 rewritten questions evaluated by GPT-4 and human for fluency, all questions were classified as coherent and fluent.
Characteristics
In the collected papers obtained from Lens, the meta-information associated with them provides subject information. Figure 2b presents the topic distribution of these papers. They cover a wide range of topics, including biochemistry, theoretical chemistry, catalysis, environmental chemistry, and material chemistry.
To delve further into the QA attributes, we performed a human analysis on a random sample of 200 examples, where we categorized the questions into three main aspects and classified the difficulty into background knowledge-required and knowledge-free categories. The three main aspects are: chemical interaction (questions about how chemicals interact or react), chemical theory (questions related to fundamental chemistry theories or principles), and chemical attributes (questions focusing on inherent properties of specific chemicals), with the majority falling under the category of chemical attributes. Examples of these are provided in Table 2. Regarding the type of reasoning required, around 71.5% of the questions require chemical knowledge for answering. The remaining questions can be addressed through semantic reasoning. For instance, the context “the metal center is really capable of back-donation to the carbene" provides the answer to the question “Back-Donation in High-Valent d0 Metal Complexes: Does It Exist?" Examples can be found in Table 2. To better illustrate the correspondence between different reasoning types and question types, we present a Sankey diagram depicted in Fig. 2c. It can be seen that different question types correspond to different reasoning types, showcasing the diversity of our dataset.
Methods
In this section, we first define the task of chemical QA, then describe our ChemMatch model in detail.
Problem Formulation
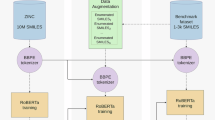
The task of building ChemMatch can be formulated as a C-class classification problem in a semi-supervised learning setting. There are labeled instances, denoted as \(\left\{{{{{\bf{q}}}}}^{s},{{{{\bf{c}}}}}^{s},{{{{\bf{y}}}}}^{s}\right\}\), and unlabeled instances, denoted as \(\left\{{{{{\bf{q}}}}}^{u},{{{{\bf{c}}}}}^{u}\right\}\), where \({{{{\bf{q}}}}}^{* },{{{{\bf{c}}}}}^{* }\in {{\mathbb{R}}}^{d}\) are the d-dimensional question and context representation, and ys is the one-hot ground-truth label. To answer a within-context question x = {q, c}, ChemMatch makes prediction \({{{{\bf{y}}}}}^{{\prime} }\) as \({{{\bf{p}}}}({{{{\bf{y}}}}}^{{\prime} }| {{{\bf{x}}}})\in {{\mathbb{R}}}^{C}\). The overview for building ChemMatch is illustrated in Fig. 3. The loss function to minimize is \({{{\mathcal{L}}}}={{{{\mathcal{L}}}}}_{s}+{{{{\mathcal{L}}}}}_{u}\). Here \({{{{\mathcal{L}}}}}_{s}\) is the supervised cross-entropy loss (\({{{\mathcal{H}}}}\)):
The unsupervised consistency loss\({{{{\mathcal{L}}}}}_{u}\) is defined by adopting a pseudo-labeling approach with consistency restriction:
where \(\hat{{{{\bf{p}}}}}\) is the pseudo-label generated for unlabeled input (see Equation (5)). \(\hat{{{{\bf{y}}}}}\) is obtained by \({{{\bf{p}}}}\left({{{\bf{y}}}}| \Omega \left({{{{\bf{x}}}}}^{u}\right)\right)\), where \(\Omega \left({{{{\bf{x}}}}}^{u}\right)\) represents the prediction based on augmented variations of the question and the context (see section 3.4). The general objective is to ensure that the predicted label of the corresponding augmented case aligns with the pseudo-labels. In this way, the vast amount of unlabeled cases is leveraged as well to optimize the prediction of y.
ChemMatch is trained using both labeled and unlabeled data. In the supervised training phase, label rebalancing is applied to adjust the loss regarding class infrequency. In the unsupervised phase, pseudo-labels are generated through pseudo-label calibration. The learning from unlabeled data is through the enforcement of consistency between the pseudo-labels and the predictions of instances augmented using SoftMix.
The minimization of both supervised loss (\({{{{\mathcal{L}}}}}_{s}\)) and unsupervised loss (\({{{{\mathcal{L}}}}}_{u}\)) is hindered by the imbalanced distribution of classes in y. Specifically, the ‘maybe’ class is significantly underrepresented compared to the ‘yes’ and ‘no’ classes. This imbalance is further aggravated when combined with an automatically annotated dataset that only includes ‘yes’ or ‘no’ labels. To address these challenges, we implement a strategy of ‘label rebalance’ during the supervised training phase and ‘pseudo-label calibration’ during the semi-supervised learning process, which are explained in detail below.
Label rebalance
From Table 1, it is evident that in the human-annotated dataset, the ‘yes’ class constitutes 65.8%, while the least represented class accounts for only 13%. Moreover, if we combine the automatically annotated dataset into training, the imbalance problem becomes even more severe, since the automatic datasets are constructed based only on ‘yes’ and ‘no’ classes. Therefore, addressing the generalization issue for the less frequent classes is crucial.
Inspired by Ref. 36, we integrate the principle of label rebalance into the traditional cross-entropy loss. Intuitively, we increase the loss weight of the less frequent class. This adaptation is advantageous for minority classes, pushing them to have broader margins and achieving higher accuracy. Let’s consider a sample labeled \({{{{\bf{y}}}}}_{i}^{s}\) which represents a class with ny training instances. The modified label-rebalanced softmax cross-entropy loss is defined as:
Here, a β value of 0 indicates that there’s no re-weighting applied. As β approaches 1, it signifies re-weighting based on the inverse of the class frequency. The hyperparameter β and the effective sample number ny allow a smooth adjustment of the class-balanced factor, ranging from no re-weighting to re-weighting by inverse class frequency.
Pseudo-label calibration
Pseudo-labels are often generated by trained models for unlabeled data37,38. By incorporating pseudo-labeled data, the model can leverage a wealth of unlabeled data, enhancing its generalization capabilities and improving prediction accuracy. To ensure the high quality of pseudo-labels, we calibrate their distribution so that it aligns with the distribution of the actual ground truth labels.
The first operation is multiplication of the predicted and ground truth distributions: This step enhances the parts of the predicted distribution that match the ground truth distribution. If a certain class has a high probability in both the predicted and ground truth distributions, its probability will further increase after multiplication. Conversely, if a class has a high probability in the predicted distribution but a low probability in the ground truth distribution, its probability will decrease after multiplication. Let \(\dot{{{{\bf{p}}}}}\in {{\mathbb{R}}}^{C}\) be the prediction of the pseudo-label of one unlabeled instance. We first multiply it with \(\bar{{{{\bf{y}}}}}\in {{\mathbb{R}}}^{C}\), which is the distribution of ground truth labels from annotated data ys.
Then, division by the past average distribution: This step aims to reduce the bias in the predicted distribution caused by the accumulation of historical data. If a certain class has appeared frequently in the past average distribution, its probability will be correspondingly reduced in the new prediction to avoid the excessive influence of past data on the current prediction. To calibrate each \(\dot{{{{\bf{p}}}}}\), we estimate its distribution in one batch \(\overline{{{{\bf{p}}}}}(y)\in {{\mathbb{R}}}^{C}\), e.g., by taking the average of the model’s predictions on unlabeled examples over the last 128 batches.
The above process can be summarized as: To adjust the predicted pseudo-labels to better reflect the true likelihood of each class, we apply the following pseudo-label calibration operation with pointwise multiplication and division:
where Normalize(a) = a/∑jaj. Together, these two steps ensure that the pseudo-labels are both accurate (by aligning with the ground truth distribution) and consistent (by forming a valid probability distribution), thereby improving the model’s ability to learn from unlabeled data.
Additionally, since ground truth labels typically adopt hard (1-hot) encoding, we further modify the calibrated pseudo-labels by applying a sharpening function:
where T is a hyperparameter. As T approaches 0, the output will approach a one-hot distribution. A reduction in T steers the model towards generating predictions with diminished entropy. Finally, we use \(\hat{{{{\bf{p}}}}}\) as the pseudo label in Equation (2) and proceed as usual with other processing.
SoftMix augmentation
To utilize the abundance of available unlabeled data and enhance the learning process, the concept of data augmentation has been extensively adopted in semi-supervised learning39. The key idea is to create data variants, make predictions, and compare them with pseudo labels to guide model training. As introduced in the semi-supervised learning framework in Section 3.1, augmenting unlabeled cases is necessary to formulate a consistency loss. Most of the existing augmentation methods are in input space. For example, augmentation on images includes rotation, cropping, and flipping, and text-domain augmentations include back translation37 and synonym substitution40. However, studies41,42 suggest that interpolations in hidden layers can capture more advanced information, enhancing semantic diversity and providing additional training signals. For example, enhancing diversity in latent spaces can improve the robustness of text generation models43. Inspired by these insights, we introduce the SoftMix augmentation operation, designed to increase diversity and strengthen robustness by latent space augmentations.
As our QA paradigm consists of the question and the input document, we naturally have two kinds of augmentation results by using back translation to translate these two parts respectively. Back translation means translating text from the source language to a target language and then translating it back to the original language. This process helps generate linguistically diverse versions of the question and document, which can improve the model’s robustness and ability to handle varied phrasings in QA tasks. We refer to the input with a back-translated question as ‘question-augmented’, and the same goes for ‘context-augmented’. Let xa be the question-augmented input representation, and xb be the answer-augmented representation. Among xa, xb and the original input xu, one can be randomly selected to act as a source of perturbation to modify the other inputs. For instance, if xa is selected for perturbation, both xu and xb are modified as:
where \({{{{{\bf{x}}}}}^{{\prime} }}^{* }\) represents the new training input derived from x* (xu and xb in this example case), and α is a hyperparameter for Beta distribution. The representations of two augmented cases are separately mixed with their own original representation to produce new training inputs with the same training target, i.e., the pseudo label. The whole process is illustrated in Fig. 3.
Note that our SoftMix operation is different from the previous RemixMatch operation in Ref. 39. In their method, they perform a weighted sum of multiple input hidden states and output states to form new training samples. In contrast, in our work, we establish a mixture operation only in the input space with representations that have similar semantic meanings, keeping the target the same. This maintains a balance between diversity in the latent space and the fundamental generative capability without interference. In the experiments section 4, we will show that our method significantly outperforms RemixMatch.
The newly generated training inputs share the same prediction objective, i.e., the pseudo label. Therefore, their predictions are compared against the pseudo label of xu, leading to the calculation of the consistency loss in Equation (2). Formally, given our augmented and mixed batches, the standard consistency loss in Equation (2) is changed to:
We additionally utilize xa, comprising a sole augmented rendition of the question and its predicted labels, excluding the application of SoftMix. This not only offers a subtle enhancement in performance but also contributes to heightened stability:
The ChemMatch model is optimized by \({{{{\mathcal{L}}}}}_{bs}+{{{{\mathcal{L}}}}}_{m}+{{{{\mathcal{L}}}}}_{c}\).
Results
Baselines
We first compare ChemMatch with a basic Supervised baseline model, which is trained by using only the human-annotated dataset. In addition, we compare ChemMatch with a biomedical baseline PubMedQA19 that leverages labeled data to produce static pseudo-labels for the unlabeled samples, which are subsequently utilized to train the classification model. PubMedQA is a multi-phase finetuning process, while our model follows end-to-end fashion with our unique softmix and rebalance operations.
We also compare with strong semi-supervised baselines:
FixMatch44 is a classic semi-supervised baseline that uses pseudo-labeling on a weakly augmented version of the data and then enforces consistency between these pseudo-labels and the predictions on a strongly-augmented version of the same data. The pseudo-label is only retained if the model produces a high-confidence prediction.
FreeMatch38 adjusts the confidence threshold of pseudo labels in a self-adaptive manner according to the model’s learning status.
SoftMatch37 derives a truncated Gaussian function to weight pseudo samples based on their confidence, which can be viewed as a soft version of the confidence threshold.
Our model differs from the above approaches by leveraging all pseudo labels and aim to enhance their accuracy.
RemixMatch39 introduces a remix operation in latent space that combines multiple cases to create new learning inputs and targets. This approach fundamentally differs from our SoftMix operation, which mixes information within a single case while maintaining the same target.
We also include open-source LLM baselines, such as Llama2-70B45, GPT-3.5, and GPT-4.
Datasets
Our ChemMatch model, though originally designed to address the imbalance phenomenon prevalent in scholarly papers, is applicable to a variety of other contexts where similar imbalances occur. Several imbalanced text classification benchmark datasets have been developed reflecting these scenarios. To evaluate our model’s effectiveness beyond the specialized chemical question answering dataset, we tested it on established benchmark classification datasets46. The AG News dataset, extracted from AG’s corpus of news articles on the web, utilizes the four largest classes from this corpus. The Yahoo Answers dataset is a topic classification dataset featuring questions and best answers from Yahoo!’s ten largest categories, including the question title, content, and best answer. The Yelp-5 dataset originates from the Yelp Dataset Challenge in 2015. We adopt the task of predicting the number of stars given by the user. Lastly, the Amazon-5 dataset from the Stanford Network Analysis Project comprises Amazon reviews. The data used for classification includes both the review title and its content.
Evaluation metrics
Each experiment is repeated five times with different data splits following previous works19,47, and we report the average test accuracy and weighted F1 scores. Accuracy reflects the proportion of accurate predictions among all instances, yet it overlooks the precision of individual classes. On the contrary, weighted-F1 computes metrics for each label and determines their average, considering the number of true instances for each label in the weighting process.
Experimental results
Outperforming similar-scale models
In Table 3, we present the performance metrics of recent baselines and our model across diverse dataset settings. The imbalance ratio γ represents how many times larger the size of the biggest class is compared to the smallest one, and γ ranges from 5 (Setting 1) to 48 (Setting 4). A few observations can be made from the table.
Firstly, semi-supervised baselines surpass the naive supervised baselines in most scenarios, which shows the necessity of learning from unlabeled cases. Secondly, it is valuable to have a larger pool of unsupervised data and supervised data. For instance, comparing Setting 2 and Setting 3, even though the supervised count remains the same, there is an improvement or consistent performance when more unsupervised data is added. Thirdly, the ChemMatch model consistently outperforms other models across all configurations. While Accuracy provides a measure of overall performance, the F1 score additionally captures the equilibrium of accuracy across various classes. Our model excels in both these metrics, thus highlighting its resilience across diverse data distributions and emphasizing its effective utilization of both supervised and unsupervised data.
Performance on general-domain datasets
The scenario of imbalanced semi-supervised learning is commonly observed in real-world settings48,49,50,51. To verify the generalizability of our model, we further evaluated our model on four benchmark datasets. To simulate an imbalanced setting, we set the imbalance ratio γ of 5 for labeled data and 150 for unlabeled data, a common setting in image classification37. For example, for AG News dataset in setting 1, the case numbers across four categories are [40, 23, 13, 8]. In setting 2, the number distribution is [200, 116, 68, 40]. The results are shown in Table 4, where our model outperforms most of the other baselines across different settings. For instance, our model achieves 87.38% accuracy with 200 labeled instances, outperforming FreeMatch’s 86.53%. These results demonstrate the generalization and robustness of our ChemMatch model in handling imbalances in different domains and settings.
Comparison with large language models
We compared our model with Llama2-70B, Meditron-70b52, GPT-3.5, GPT-4 across 200 sampled cases, where the chain-of-thought prompt is in the Supplementary Note 1. The accuracy and F1 results are shown in Fig. 4. Our model surpasses the three baseline models probably because it is trained explicitly on the chemical corpus, hence, it’s enriched with corresponding knowledge. The advantages of our model become more evident when considering the large size and great computational source of LLMs. Additionally, Meditron-70b fails to provide answers to the questions and instead simply repeats them, as observed in related queries at https://github.com/epfLLM/meditron/issues/13, demonstrating that further training is required for this model.
Prompt discussion
We employed various strategies when testing LLMs, including chain-of-thoughts and few-shot learning, in designing the prompts. However, we observed that neither strategy substantially enhanced performance. For few-shot learning, the limited improvement may be attributed to the dissimilarity in content among the test questions, indicating a necessity for more targeted selection of in-context learning cases. We assume the reason is that the limitations of LLMs in chemical QA task are more related to a deficit in domain-specific scientific knowledge rather than the thinking strategy. This insight directs us towards strengthening LLMs’ domain-specific scientific knowledge in chemical QA tasks.
Case study
We also give an error analysis on the output of LLMs in Fig. 5. Generally, we observed that both GPT-3.5 and GPT-4 often provide ambiguous ‘maybe’ answers, even when the input clearly warrants a definitive ‘yes’ or ‘no’ response, For instance, the conclusion is initially presented in the input as ‘the proton can be encapsulated’. The subsequent details then delve into the specific conditions under which the proton can or cannot be encapsulated, creating confusion for the LLMs, as evidenced by the outlined reasons. These examples highlight the inconsistency between the LLM’s reasoning process and its final conclusions, which points to further improvement.
DISCUSSION
Ablation study
In Table 5, we assess the contributions of ChemMatch’s main components in four Settings. Take setting 4 as an example, the full ChemMatch model, with an accuracy of 74.28% and F1 score of 71.06%, outperforms its variants. Excluding label rebalancing results in reduced performance, with accuracy and F1 scores dropping to 73.96% and 70.32%, respectively. The performance drops even further when pseudo-label calibration is removed. These findings underscore the importance of class balancing. Finally, the lack of the SoftMix component (when \({{{{\mathcal{L}}}}}_{m}\) is absent and only \({{{{\mathcal{L}}}}}_{c}\) is utilized) hurts performance in both metrics, underscoring the benefits of augmenting diversity. The comparative performance is consistent throughout different datasets, which demonstrates the robust effectiveness of our proposed modules.
Effectiveness of label rebalance
We next analyze the three components in detail, starting with a simple numerical study. The first component is label rebalancing. We examine the influence of the size of β on the class-based weight, as shown in Equation (3). When β = 0, it corresponds to no re-weighting, and as β approaches 1, it corresponds to re-weighting by inverse class frequency. The proposed concept of the effective number of samples enables us to use the hyperparameter β to smoothly adjust the class-balanced term between no re-weighting and re-weighting by inverse class frequency. In Fig. 6b, we demonstrate that the class-balanced term always improves the performance of the original loss, and larger values of β yield more significant performance gains.
Effectiveness of Pseudo-label calibration
In Section 3.3, we introduce two steps to align the prediction distribution for unlabeled cases with the ground truth distribution. Herein, we present the histogram distribution of the ground truth labels, the predictions from the baseline FixMatch, and our model ChemMatch in Fig. 7. It can be observed that FixMatch has significantly fewer predictions for the minority class, while our ChemMatch produces a prediction distribution similar to the ground truth.
We also calculate the KL divergence53 between predictions and ground truth labels. The KL loss between FixMatch and the labels is 0.0467, while it is 0.0028 for ChemMatch. This indicates that the distribution of Pred2 is closer to the target distribution compared to Pred1, as reflected by the lower KL divergence value. This demonstrates that our techniques for pseudo-label calibration are effective, and the predicted labels are of good quality, closely resembling the ground truth labels. This provides a favorable condition for semi-supervised learning.
Effectiveness of SoftMix operation
Apart from the numerical study, we conduct visualizations to provide a more intuitive understanding of the proposed structure. Firstly, we demonstrate the effectiveness of the SoftMix operation through t-SNE projection. In Supplementary Fig. S1, we project the original labeled text, the augmented text using back translation, and the hidden vector obtained by the SoftMix operation, respectively. It can be seen that the augmented text is close to the original text, which means that the back translation operation brings limited diversity to the training corpus. Then, for the gray nodes projected by the hidden cases generated by the SoftMix operation, they are farther away from both the original document and the augmentation in the input space, and are more dispersed in the latent space. This indicates that the SoftMix operation can generate more diverse and informative representations compared with the input space augmentation, potentially leading to improved model performance.
Comparison with RemixMatch baseline
We also tried an alternative remix operation proposed by Ref. 39. It introduces a remix operation in latent space that combines multiple cases to create new learning inputs and targets. This approach differs from our SoftMix operation, which mixes information within a single case while maintaining the same target. As shown in Table 5, with the remixmatch component the model performs poorly on textual tasks. This observation aligns with the previous findings at: https://github.com/microsoft/Semi-supervised-learning/blob/main/results/usb_nlp.csv. A plausible explanation is that, in the text domain, the semantic vector representations cannot be mixed similarly to the computer vision domain, which can cause confusion in the training process and interfere with performance.
Comparison with PubMedQA baseline
PubMedQA is a model proposed by PubMedQA19. One significant difference is that it focuses on the biomedical domain, whereas the performance of language models in the chemistry domain remains largely unexplored. In terms of data collection, our work encompasses papers from various sources such as Elsevier and Springer, which are diverse and cover multiple topics. Regarding model design, PubMedQA undergoes a multi-phase training process using either ground truth labels or pseudo labels. In contrast, our model follows an end-to-end approach, where it is trained simultaneously with both labeled and pseudo labels. We also introduce a module that includes a softmix augmentation operation to more effectively utilize imbalanced datasets, which significantly outperforms PubMedQA.
Conclusion and broader impacts
In this study, we introduce the a large-scale chemical question-answering dataset, gathered from academic sources. Given the inherent imbalance of the data attributes, we further present ChemMatch, a question-answering model specifically adapted for imbalanced semi-supervised learning. This model introduces label-rebalance and pseudo-calibration operations to address the imbalance issue. Experimental results show that ChemMatch surpasses recent classification baselines and LLMs. Our dataset holds the potential for additional scientific investigation. For instance, it can be used for testing domain-specific language models on their understanding of complex chemical concepts. It can also examine chemical research information retrieval systems, particularly in matching questions with corresponding documents.
Data availability
The code and data sample is publicly available at https://github.com/iriscxy/chemmatch. Our dataset is drawn from various academic platforms, each having distinct data protection policies. A significant portion of our dataset is sourced from the lens.org34 website, a platform that actively promotes the distribution and sharing of data. As per the guidelines detailed at https://about.lens.org/policies/#attribution, we are allowed to release the dataset with the Lens ID. The extensive scale of 26,000 cases holds significant potential and is expected to provide considerable benefits to the community. As for data sourced from other sites like Elsevier, which maintains strict data usage policies at https://www.elsevier.com/about/policies/copyright/permissions, we release the DOI of the files within our dataset alongside our data collection code. This approach enables users to recollect our data collection steps, but always within the constraints set by the original data providers’ permissions.
Code availability
The code and data sample is publicly available at https://github.com/iriscxy/chemmatch.
References
Hu, S., Zou, L., Yu, J. X., Wang, H. & Zhao, D. Answering natural language questions by subgraph matching over knowledge graphs. IEEE Trans. Knowl. Data Eng. 30, 824–837 (2017).
Lan, Y. et al. Complex knowledge base question answering: A survey. IEEE Trans. Knowl. Data Eng. 35, 11196–11215 (2022).
Christmann, P., Saha Roy, R. & Weikum, G. Conversational question answering on heterogeneous sources. In Proceeding of International Conference on Research on Development in Information Retrieval (2022).
Qu, C. et al. Open-retrieval conversational question answering. In Proceeding of International Conference on Research on Development in Information Retrieval (2020).
Auer, S. et al. The sciqa scientific question answering benchmark for scholarly knowledge. Sci. Rep. 13, 7240 (2023).
Zheng, S., Li, Y., Chen, S., Xu, J. & Yang, Y. Predicting drug–protein interaction using quasi-visual question answering system. Nat. Mach. Intell. 2, 134–140 (2020).
Jin, Q. et al. Hidden flaws behind expert-level accuracy of multimodal gpt-4 vision in medicine. ArXiv (2024).
Maharjan, J. et al. Openmedlm: prompt engineering can out-perform fine-tuning in medical question-answering with open-source large language models. Sci. Rep. 14, 14156 (2024).
Mahbub, M. et al. Question-answering system extracts information on injection drug use from clinical notes. Commun. Med. 4, 61 (2024).
Clark, C. et al. Boolq: Exploring the surprising difficulty of natural yes/no questions. In Proceeding of of North American Chapter of the Association for Computational Linguistics (2019).
Wadden, D. et al. Fact or fiction: Verifying scientific claims. In Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP), 7534–7550 (2020).
Wang, L. L. Using machine learning to verify scientific claims (2023).
Ghoshal, A. et al. Quaser: Question answering with scalable extractive rationalization. In Proceeding of International Conference on Research on Development in Information Retrieval (2022).
Garcia-Silva, A. et al. Spaceqa: Answering questions about the design of space missions and space craft concepts. In Proceeding of International Conference on Research on Development in Information Retrieval (2022).
Peretz, G., Arraf, M. & Radinsky, K. What if: Generating code to answer simulation questions in chemistry texts. In Proceedings of the 46th International ACM SIGIR Conference on Research and Development in Information Retrieval, 1335–1344 (2023).
Goldsmith, E. J., Mendiratta, S., Akella, R. & Dahlgren, K. Natural language query in the biochemistry and molecular biology domains based on cognition search™. Summit Transl. Bioinforma. 2009, 32 (2009).
Krithara, A., Nentidis, A., Bougiatiotis, K. & Paliouras, G. Bioasq-qa: A manually curated corpus for biomedical question answering. Sci. Data 10, 170 (2023).
Jablonka, K. M., Schwaller, P., Ortega-Guerrero, A. & Smit, B. Leveraging large language models for predictive chemistry. Nat. Mach. Intell. 6, 161–169 (2024).
Jin, Q., Dhingra, B., Liu, Z., Cohen, W. & Lu, X. Pubmedqa: A dataset for biomedical research question answering. In Proceeding of Empirical Methods in Natural Language Processing (2019).
Jin, D. et al. What disease does this patient have? a large-scale open domain question answering dataset from medical exams. Appl. Sci. 11, 6421 (2021).
Wang, H., Zhou, L., Zhang, W. & Wang, X. Literatureqa: A qestion answering corpus with graph knowledge on academic literature. In Proceeding of CIKM (2021).
Laurent, J. M. et al. Lab-bench: Measuring capabilities of language models for biology research. arXiv preprint arXiv:2407.10362 (2024).
Pan, J. Large language model for molecular chemistry. Nat. Comput. Sci. 3, 5–5 (2023).
Tibo, A., He, J., Janet, J. P., Nittinger, E. & Engkvist, O. Exhaustive local chemical space exploration using a transformer model. Nat. Commun. 15, 7315 (2024).
M. Bran, A. et al. Augmenting large language models with chemistry tools. Nature Machine Intelligence 1–11 (2024).
Oniani, D. et al. Emerging opportunities of using large language models for translation between drug molecules and indications. Sci. Rep. 14, 10738 (2024).
Wei, Z. et al. Chemistryqa: A complex question answering dataset from chemistry (2020).
Zhou, X., Nurkowski, D., Mosbach, S., Akroyd, J. & Kraft, M. Question answering system for chemistry. J. Chem. Inf. Model. 61, 3868–3880 (2021).
Mirza, A. et al. Are large language models superhuman chemists? arXiv preprint arXiv:2404.01475 (2024).
M. Bran, A. et al. Augmenting large language models with chemistry tools. Nat. Mach. Intell. 6, 525–535 (2024).
Guo, T. et al. What can large language models do in chemistry? a comprehensive benchmark on eight tasks. Adv. Neural Inf. Process Syst. 36, 59662–59688 (2023).
Lu, P. et al. Learn to explain: Multimodal reasoning via thought chains for science question answering. Adv. Neural Inf. Process. Syst. 35, 2507–2521 (2022).
Huang, C., Li, Y., Loy, C. C. & Tang, X. Learning deep representation for imbalanced classification. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 5375–5384 (2016).
Jefferson, O. A. et al. Mapping the global influence of published research on industry and innovation. Nat. Biotechnol. 36, 31–39 (2018).
Toutanvoa, K. & Manning, C. D. Enriching the knowledge sources used in a maximum entropy part-of-speech tagger. In Proceeding of Empirical Methods in Natural Language Processing (2000).
Cui, Y., Jia, M., Lin, T.-Y., Song, Y. & Belongie, S. Class-balanced loss based on effective number of samples. In Proceeding of International Conference on Computer Vision and Pattern Recognition (2019).
Chen, H. et al. Softmatch: Addressing the quantity-quality tradeoff in semi-supervised learning. In Proceeding of International Conference on Learning Representations (2023).
Wang, Y. et al. Freematch: Self-adaptive thresholding for semi-supervised learning. Proceeding of International Conference on Learning Representations (2023).
Berthelot, D. et al. Remixmatch: Semi-supervised learning with distribution alignment and augmentation anchoring. Proceeding of International Conference on Learning Representations (2020).
Gan, Y. et al. Towards robustness of text-to-sql models against synonym substitution. In Proceeding of Association for Computational Linguistics, 2505–2515 (2021).
Zeiler, M. D. & Fergus, R. Visualizing and understanding convolutional networks. In Proceeding of ECCV (2014).
Verma, V. et al. Manifold mixup: Better representations by interpolating hidden states. In Proceeding of International Conference on Machine Learning (2019).
Chen, X. et al. Improving the robustness of summarization systems with dual augmentation. Proceeding of Association for Computational Linguistics (2023).
Sohn, K. et al. Fixmatch: Simplifying semi-supervised learning with consistency and confidence. Proc. Neural Inf. Process. Syst. 33, 596–608 (2020).
Touvron, H. et al. Llama 2: Open foundation and fine-tuned chat models. arXiv preprint arXiv:2307.09288 (2023).
Zhang, X., Zhao, J. & LeCun, Y. Character-level convolutional networks for text classification. Proceeding of Neural Information Processing Systems (2015).
Lin, M. et al. Improving model fairness in image-based computer-aided diagnosis. Nat. Commun. 14, 6261 (2023).
Tzaban, H. et al. Product bundle identification using semi-supervised learning. In Proceeding of International Conference on Research on Development in Information Retrieval (2020).
Kim, J. et al. Distribution aligning refinery of pseudo-label for imbalanced semi-supervised learning. Proc. Neural Inf. Process. Syst. 33, 146567–14579 (2020).
Lee, H., Shin, S. & Kim, H. Abc: Auxiliary balanced classifier for class-imbalanced semi-supervised learning. Proc. Neural Inf. Process. Syst. 34, 7082–7094 (2021).
Wei, C., Sohn, K., Mellina, C., Yuille, A. & Yang, F. Crest: A class-rebalancing self-training framework for imbalanced semi-supervised learning. In Proceeding of International Conference on Computer Vision and Pattern Recognition (2021).
Chen, Z. et al. Meditron-70b: Scaling medical pretraining for large language models. arXiv preprint arXiv:2311.16079 (2023).
Hershey, J. R. & Olsen, P. A. Approximating the Kullback Leibler divergence between Gaussian mixture models. In 2007 IEEE International Conference on Acoustics, Speech and Signal Processing-ICASSP’07, vol. 4, IV–317 (IEEE, 2007).
Author information
Authors and Affiliations
Contributions
Xiuying Chen and Tairan Wang conceived the ideas, conducted the experiments, and co-wrote the paper. Taicheng Guo, Kehan Guo, Juexiao Zhou, and Haoyang Li collected and labeled the dataset. Zirui Song performed additional experiments during the rebuttal phase. Xin Gao and Xiangliang Zhang contributed to refining and polishing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Chemistry thanks Sutanay Choudhury, Qingyun Wang, and the other, anonymous, reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, X., Wang, T., Guo, T. et al. Unveiling the power of language models in chemical research question answering. Commun Chem 8, 4 (2025). https://doi.org/10.1038/s42004-024-01394-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42004-024-01394-x
This article is cited by
-
Evaluating and mitigating bias in AI-based medical text generation
Nature Computational Science (2025)