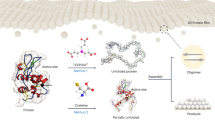
Abstract
Deep eutectic solvents (DESs) have emerged as powerful environments to enhance enzymatic reactions, formulate therapeutic proteins, and develop protein-based biomaterials. Despite the wide range of properties that could be achievable through the compositional design of DESs, protein solubilization only happens in a relatively narrow range of hydrophilic DESs. Here, we use surface-modification for the generalized solubilization of proteins in both hydrophilic and hydrophobic DESs. Using surface-modified myoglobin as a model, we show that both DES polarity and hydrogen bond capacity play important roles in dictating the conformational state of the protein. In the hydrophilic DES the protein displays a near-native conformation with an improvement of the thermal stability of + 28 °C compared to aqueous solutions. In contrast, hydrophobic DESs stabilize partially folded intermediates which can refold from temperatures as high as 190 °C. As such, our approach provides a platform to generalize protein incorporation into anhydrous DESs that could be exploited in biocatalysis, biomolecule stabilization, and biomaterials.

Similar content being viewed by others
Introduction
Controlling the structure and function of proteins is critical for the development of biomaterials and expanding the repertoire of enzyme-based biocatalysis. In these regards, deep eutectic solvents (DESs) are potentially powerful tools for modulating enzyme activity and biomolecule architecture1. DESs are non-aqueous solvents typically obtained through the mixture of a hydrogen bond acceptor (HBA, e.g., choline chloride) and a hydrogen bond donor (HBD, e.g., glycerol). When mixed at a specific molar ratio (1:2 choline chloride:glycerol), these components elicit a depression in the melting point of the mixture beyond that of the ideal mixture of the precursors, as governed by hydrogen bonding, electrostatics, and van der Waals interactions, yielding a room-temperature liquid phase2,3. The most attractive characteristic of DESs is the possibility of formulating them through a virtually infinite number of combinations of precursors, which results in an extremely wide variety of solvents with varied physiochemical properties (e.g., hydrophobicity, solvent cohesivity, and charge density) that allow these “designer” solvents to be tailored for a specific application4,5,6. Recently, DESs have been shown to support protein folding7,8, enzymatic catalysis9,10, biomolecule stabilization11,12, protein crystallization13, and to support the development of protein-based eutectogels14,15,16. These developments are all linked by the fine control of protein conformational state as dictated by both specific solvent-protein and non-specific interactions of the solvation matrix around the protein17,18. Furthermore, DESs provide unique opportunities for biocatalysis through the solubilization of otherwise recalcitrant molecules19,20, coupled with the relative ease of recycling the DES-biocatalyst system21.
Although there has been a rapid evolution in the field, current research is limited by biomolecule solubility in DESs. This is particularly obvious for hydrophobic DESs, since no evidence of protein stabilization has been reported thus far. While hydrophilic DESs constitute a powerful platform, the inclusion of hydrophobic DESs would greatly expand the application prospects of these solvents for biomaterial design and biocatalysis22. For example, alternative enzymatic routes could be explored by finding a media for the solubilization of aqueous insoluble substrates or even inducing enzyme promiscuity toward specific substrates19,23. Additionally, the solubilization of hydrophobic pharmaceutical compounds could be exploited for drug delivery applications15,24. Protein engineering, such as directed evolution and mutagenesis, has become the benchmark method to tailor protein stability and function in DESs25,26. However, these methods are typically costly with respect to time and instrumentation required, and do not provide a general pathway for DES incorporation. In contrast, facile surface-engineering of proteins with a polymer-surfactant corona is a versatile method for protein solubility and stabilization in a broad range of non-aqueous solvents27,28,29. In particular, surface-modified proteins have been effectively incorporated into anhydrous ionic liquids, allowing to exploit enzymatic activity in unconventional media23,28,30, and potentially showing the suitability of this approach for the generalized solubilization of proteins in DESs.
Here, we explore the structure and stability of surface-modified myoglobin solubilized in a series of compositionally varied DESs. Previous studies demonstrated that protein modification was required to solubilize proteins in anhydrous ionic liquids, where the protein retain a globular structure regardless the polarity of the solvent28. In contrast, DESs introduce the possibility of accessing different conformational states in acute response to solvent composition. In doing so, the compositional design of DESs impacts protein structure and stability in a highly nuanced way that depends on both solvent polarity and hydrogen bonding capabilities. Notably, hydrophobic solvents allow for a near fully recoverable secondary structure, even after near complete disordering at temperatures as high as 190 °C. As such, we show that DESs are a powerful platform for the fine control of protein conformation and stability. In particular, we demonstrate that protein surface-modification allows for generalized solubilization, potentially informing further research in the deployment of DESs for the design of biomaterials and as a key component of future biocatalysis engineering.
Results and discussion
Myoglobin was chosen as a well-characterized archetype for this investigation, as its structural response to changes in the chemical environment is well known31,32,33. The protein was surface-modified using established techniques to yield the protein-polymer surfactant nanoconjugate [C-Mb][S]27. To investigate the effect of DES composition on protein structure and stability, [C-Mb][S] was dissolved in three solvents of varying hydrophobicity and hydrogen bond capabilities as determined from the Kamlet–Taft parameters (Table 1, Supplementary Fig. 1), namely: 1:2 choline chloride:glycerol (ChCl:Glyc)34, 1:4 tetrabutylammonium chloride:glycerol (TBAC:Glyc)35, and 1:2 tetrabutylammonium chloride:octanoic acid (TBAC:OA)36. All solutions were homogeneous and isotropic in appearance, confirming full incorporation of the nanoconjugate in the three anhydrous DESs.
Far-UV synchrotron radiation circular dichroism (SRCD) was used to determine the impact of the DESs on the secondary structure of the solvated myoglobin nanoconstruct (Fig. 1a). Overall, SRCD data showed subtle changes in myoglobin secondary structure as compared to the aqueous control, where spectra deconvolution using BeStSel revealed the differences between the solvents (Table 2)37. The secondary structure of [C−Mb][S] in ChCl:Glyc was estimated at being broadly the same to that in aqueous conditions, only differentiated by a slight decrease of the α-helix content that was concomitant with an increase of the β-sheet content. In contrast, replacing the hydrogen bond acceptor to TBAC caused measurable changes to the local folding of myoglobin, with a decrease in the α-helix in favor of more unordered secondary structure content. In the most hydrophobic DES tested, TBAC:OA, the helicity of [C-Mb][S] significantly increased compared to that in TBAC:Glyc.
The protein nanoconstruct was dissolved in H2O (gray squares), ChCl:Glyc (red triangles), TBAC:Glyc (blue circles), and TBAC:OA (green diamonds): a SRCD spectra, b SANS data and fits, and c UV-vis absorbance normalized to absorbance at 280 nm. For SANS characterization the solvents were deuterated, and isotopic labeling did not cause any significant change in protein behavior (Supplementary Note 2). The fits in (b) are presented for the entire q-range (black solid lines) and for the high q-expansion of the data (dashed lines). Data and models in (b) have been offset for clarity. Where not seen, error bars are within the markers. All data were recorded at 25 °C.
In light of the impact on secondary structure, we used small angle neutron scattering (SANS) and UV-vis to investigate the effect of DES composition on the tertiary structure of the protein (Fig. 1b, c). In agreement with SRCD, the SANS analysis for [C-Mb][S] in ChCl:Glyc confirms a preserved globular conformation of the protein with similar dimensions to that in aqueous solution (Fig. 1b, Table 2)38. In fact, the SANS signal attributed to the nanoconstruct folding (Supplementary Note 3) displayed a similar shape in ChCl:Glyc, aqueous buffer, and 150 mM NaCl aqueous solution upon deconvolution of interparticle interactions. This indicated that the form factor and, thus, the conformation of the protein nanoconstruct was similar in these solvents. This observation was confirmed by UV-vis (Fig. 1c), which showed a Soret absorption band centered at 409 nm, indicative of maintained tertiary structure that was equivalent to that observed in aqueous solution. However, the interactions between protein nanoconstructs in solution changed upon solvent replacement. Unlike in aqueous buffer, supramolecular clustering of the globular nanoconstructs in ChCl:Glyc was observed in the SANS signal at low q (q < 0.06 Å-1, Porod slope = 2.4 ± 0.2), possibly associated to the screened electrostatics in the DES38. Although no aggregation was observed during sample preparation or characterization, it should be noted that samples were centrifuged at 13200 rpm for 10 min to remove any possible aggregates. Thus, the signals arising at low q of the SANS data are attributed to the formation of [C-Mb][S] clusters. The SANS results of [C-Mb][S] in TBAC:Glyc showed a shift in tertiary structure that was also in agreement with the corresponding SRCD spectra. Specifically, there was a slight reduction in the minor radius to 18.0 Å and a 1.7-fold increase in the major radius of the protein (Table 2). This suggested the stabilization of a compact molten globule, possibly mediated by hydrophobic and electrostatic interactions39, where the native tertiary structure of the protein nanoconjugate was partially disrupted with the associated impact on the secondary structure. Again, this was confirmed with UV-vis, which showed a main Soret band at 409 nm with the appearance of an intense shoulder at ca. 385 nm (Fig. 1c), attributed to partially unfolded states present in this DES, and consistent with a molten globule architecture31. This shift in the structure of [C-Mb][S] in TBAC:Glyc aligns with the previously observed tendency of DESs to partially disrupt the ordered regions of certain proteins, such as BSA, often associated with specific solvent-protein interactions17,18.
In contrast, particularly given to the highly ordered secondary structure observed by SRCD, the SANS results for [C-Mb][S] in TBAC:OA indicated a significant shift in the overall morphology with a reduction in the minor radius to 11.3 Å and a dramatic 5.7-fold increase in the major radius (Table 2). This suggested a near total loss of native tertiary structure, but with preserved local folding as suggested by CD analysis. In fact, the experimental cross-sectional size is similar to that of isolated secondary structure motifs (e.g., ca. 6 Å for an isolated α-helix)40, and the low-q scattering showed the characteristic feature of an entangled network of unfolded protein domains (Porod slope = 1.8 ± 0.2)41. UV-vis confirmed the disruption of tertiary structure with a broad Soret band now centered at 385 nm (Fig. 1c), indicative of major changes in the heme binding pocket31. We hypothesize that the amphipathic nature of the hydrogen bond donor (octanoic acid) in this DES is behaving in a similar manner to surfactant denaturants, attacking the hydrophobic pockets of the myoglobin and causing a loss of the globular structure33,39. However, this solvent promotes the ordering of the secondary structure into helical motifs, as observed for peptides in the presence of ionic amphiphiles42.
These results showed that DES composition greatly impacted both secondary and tertiary structure of the solubilized protein. To determine the origin of these changes, Kamlet-Taft parameters for these solvents were determined using standard spectroscopic assays (Table 1)43,44. Both ChCl:Glyc and TBAC:Glyc had comparable polarities (π*) but differed in hydrogen bond acidity and basicity (α and β respectively). Given the ability of these two solvents to direct a certain degree of [C-Mb][S] folding, changes in the secondary and tertiary structure of [C-Mb][S] could be attributed to the differences in hydrogen bonding capabilities of the solvents. Specifically, the increase in hydrogen bond basicity for TBAC:Glyc may indicate a greater ability to disrupt hydrogen bonding within the protein, which could cause the partial unfolding of the protein secondary structure and, concomitantly, the overall conformation16. Conversely, the lower polarity of TBAC:OA, as expected from the large hydrophobic domain of the fatty acid36, dominates the interaction between protein and solvent, thus causing an increase in the helicity and the unfolding of the tertiary structure of the protein. These findings contrast with the behavior of [C-Mb][S] in ionic liquids, as the protein nanoconstruct displayed similar structure despite varying the properties of the solvents28.
Having observed how DES composition affected the structure of the solubilized protein, we turned to temperature-dependent measurements to probe this further and examine any relationship to protein stability. Previous investigations showed that the modified proteins become more thermally resilient when solubilized in ionic liquids23. Temperature-dependent SRCD confirmed that this was also the case for DESs. The SRCD data display a gradual decrease in the ordered secondary structure of [C-Mb][S] with increasing temperature, as expected from the thermal denaturation of the protein secondary structure (Fig. 2). In DESs, denaturation proceeded analogously to aqueous environments conforming to a 2-state denaturation, but over a much extended temperature range than in water (Fig. 2d)28.
a ChCl:Glyc, (b) TBAC:Glyc, (c) TBAC:OA, and (d) aqueous buffer, showing progressive thermal denaturation between 0 °C (blue) and 200 °C (red) in DESs and between 20 °C (blue) and 90 °C (red) in aqueous buffer. All data were recorded in 5 °C steps. e Plots of fraction denatured against temperature and (f) Gibbs free energy of denaturation as a function of temperature over the linear transition region of [C−Mb][S] in DESs. These parameters were calculated from SRCD data for [C-Mb][S] in ChCl:Glyc (red triangles), TBAC:Glyc (blue circles), TBAC:OA (green diamonds), and water (black squares). Data in were fitted to the models (dotted lines) to determine the thermodynamics of protein denaturation as described in the Supplementary Methods.
As anticipated, the half denaturation temperature (Tm) of [C-Mb][S] in DESs was higher than in aqueous solution (Table 3, Fig. 2e). The greatest increase in Tm was observed in ChCl:Glyc, with a 27.6 °C improvement in the thermal stability of the protein. Reflecting the observed differences in structure (Fig. 1), the stability of the protein in the hydrophobic DESs was lower than in ChCl:Glyc. The loss of folding resulted in a decrease in the thermal stability of the protein, with the partially folded states in TBAC:Glyc displaying a Tm of 74.7 °C and the unfolded conformations in TBAC:OA melting at 68.6 °C. Despite the lower Tm, the secondary structure of [C-Mb][S] in the hydrophobic DESs seems to be more temperature resilient, as observed in the SRCD signal, compared to that in ChCl:Glyc, which almost vanished at high temperature. In fact, the overall conformation in TBAC-based DESs remains practically unchanged at 75 °C and 125 °C, while [C-Mb][S] in ChCl:Glyc unfolds and aggregates at high temperature (Supplementary Note 4).
Thermodynamic analysis (Table 3, Fig. 2f) suggested that the relative stabilization in DESs was driven by a reduction in the entropy of denaturation (ΔSm) compared to aqueous solutions, offsetting the apparent reduction in the enthalpic (ΔHm) barrier. This is possibly attributed to the stabilization of a conformationally flexible structure in DESs that can gradually adapt to heating, consistent with a shift in hydrogen bond and polarity patterns between these two solvents. The results suggest that the greater hydrogen bond basicity and lower hydrogen bond acidity of TBAC:Glyc was destabilizing the structure of the protein. Similarly, the lower polarity and amphipathic character of TBAC:Glyc further perturbs the cohesivity of the intra-protein interactions that controls and maintains folding16.
Previous work showed that [C-Mb][S] was able to refold from high temperatures upon cooling in anhydrous conditions27. In a bid to elucidate a greater understanding of the potential mechanism to DES-dependent structuration of [C-Mb][S], we therefore turned to SRCD and SANS refolding studies (Fig. 3). In ChCl:Glyc, SRCD showed that once in the denatured state (190 °C), [C-Mb][S] did not recover any structure once returned to ambient temperatures (Fig. 3a). SANS revealed that this was likely the result of a non-specific aggregation process, as shown by the major increase of the signal at q < 0.04 Å-1, that occurred at temperatures around the Tm (Fig. 3d). Conversely, in TBAC:Glyc [C-Mb][S] was able to recover a significant amount (76%) of secondary structure after being fully denatured at 190 °C (Fig. 3b). Temperature-dependent SANS profiles indicated that this was also reflected in the global conformation of the protein (Fig. 3e), as our results show the unfolding of the protein at 125 °C (req = 10.6 ± 0.7 Å, rpo = 872 ± 127 Å), with a near complete return of dimensions from above the denaturation temperature (req = 19.4 ± 0.6 Å, rpo = 95.8 ± 4.2 Å, Table S1). Likewise, the protein’s ambient secondary structure was well-preserved for [C-Mb][S] in TBAC:OA (87%) (Fig. 3c). Again, this return of structure was also observed in the SANS profiles, where initial conformation is completely regained with no evidence of temperature-induced degradation (Fig. 3f).
a–c Far-UV SRCD of [C-Mb][S] dissolved in ChCl:Glyc (a), TBAC:Glyc (b), TBAC:OA (c), showing spectra at 30 °C (blue circles), 190 °C (red triangles), and subsequent recovery after returning to 30 °C (green squares). SANS profiles for [C-Mb][S] dissolved in d9-ChCl:d8-Glyc (d), d36-TBAC:d8-Glyc (e), and d36-TBAC:d16-OA (f) showing temperature dependence at 25 °C (blue circles), 125 °C (red triangles) and upon returning to 25 °C (green squares). The fits are presented for the entire q-range (black solid lines) and for the high q-expansion of the data (dashed lines). Data and models in (d), (e), and (f) have been offset for clarity. Where not seen, error bars are within the markers.
These refolding studies demonstrate that protein structure and stability are intrinsically linked to DES composition. ChCl:Glyc was able to maintain aqueous-like secondary and tertiary structure and provide the necessary environment for robust thermal stability. However, protein denaturation occurred via irreversible formation of aggregates. In comparison, whilst TBAC:Glyc caused some structural instabilities in the protein that resulted in a subtle loss of ordered secondary structure, high levels of refolding were possible from very high temperatures (190 °C). Kamlet-Taft parameters suggest that ChCl:Glyc and TBAC:Glyc differ mainly in the hydrogen bond ability of the solvent. As such, the TBAC:Glyc hydrogen bonding capabilities dictate the destabilization of a partially folded state of the protein by favoring protein-solvent interactions, which in turn provide a thermodynamic basis for refolding through a thermally resilient conformation. Interestingly, significant refolding ensues a corresponding destabilization of the folded state, where perhaps the lower hydrogen bond acidity means unsatisfied hydrogen bonding in the denatured state. The greater refolding capability observed in TBAC:OA supported this hypothesis. Specifically, TBAC:OA has a lower hydrogen bond acidity and polarity than TBAC:Glyc, and refolding capability increased accordingly.
Taken together, our results indicate an intimate relationship between protein structure and DES composition. Previous investigations have demonstrated that interactions with DESs, mainly electrostatic45,46, hydrophobic47,48, and hydrogen bond49,50, dictate the mesoscopic structure of macromolecules. Protein secondary structure retention appears to be linked to hydrogen bond acidity and basicity of the solvent, whereas protein tertiary structure is controlled by solvent polarity. As previously observed for polymer solvation in DESs, the weaker interaction of ChCl:Glyc allows the protein to fold into a near-native conformation, while an increase in basicity displaces intra-protein interactions and causes its partial unfolding50. In contrast, TBAC:OA disrupts the global structure of [C-Mb][S], which could be attributed to the amphipathic nature of the DES constituents, as it has been shown that solvotropes can penetrate the hydrophobic domains of macromolecules and alter their overall structure in DESs47,48. Intriguingly, there also appears to be a balance that could be shifted whereby hydrogen bond capabilities of the solvent could be tuned to enhance refolding of the protein. It is clear that this phenomenon cannot be simply understood in terms of isolated interactions between the DES constituents with the protein, and further studies are required to understand this effect. Regardless, our work indicates that DESs generally provide a suitable environment for high thermal stability, can support renaturation of completely disrupted structures, and provide a unique opportunity to fine-tune the global architecture of the protein.
Conclusions
In conclusion, we have demonstrated that DES composition acutely affects the structure and stability of solubilized proteins. Whilst the thermal stability of proteins in DESs is greatly improved, both the hydrogen bond capability and the polarity of the solvent have different effects on the protein’s secondary structure, tertiary structure, collective behavior, and ability to refold from a temperature-induced denatured state. By and large, our results indicate that hydrogen bond capabilities dictate secondary structure, whilst polarity mainly controls tertiary structure. Therefore, our results showcase DESs as highly tunable environments for the fine control of protein behavior. Furthermore, DESs —provided adequate composition— allow for refolding from high temperatures (190 °C). As such, DESs are capable of maintaining thermal resilience of the protein nanoconjugates as well as modulating the reversibility of unfolding processes through the design of solvent composition. Notably, these are seemingly connected to the conformational state of the protein, ultimately dictated by solvent composition.
These observations contrast with previous investigations of proteins in ionic liquids where structure and stability were mostly independent of solvent composition28. As such, combining surface engineering of proteins with DES composition holds the potential to provide a platform for developing new technologies involving these systems. For instance, folded conformations of therapeutic proteins or enzymatically active proteins could be exploited in non-aqueous biomolecular preservation or biocatalysis respectively11,23. In addition, the great variety of conformational states that can be achieved through the compositional design of DESs could be exploited in the development of solvent-controlled biomaterials, where partially folded and unfolded conformations lead to protein-based materials with designed mechanical properties16. The results presented here clearly only scratch the surface of possible compositions and their impact on protein structure and function. As such, our future studies will delve into the behavior of protein nanoconjugates in a broad range of compositionally designed DESs such as non-amphipathic hydrophobic DESs, ternary mixtures, and hydrated systems22,49. Altogether, we expect this synthetically-accessible approach will expedite the use of a wider range of DESs as protein environments for a wide range of applications.
Methods
Surface modification of myoglobin
Myoglobin (Mb) was cationized using N,N’-bis(2- aminoethyl)-1,3-propanediamine (C−Mb) and combined with glycolic acid ethoxylate lauryl ether (S) to yield the protein nanoconstruct [C−Mb][S]23,27. The resulting product was isolated by filtration and centrifugation using 10,000 MWCO centrifugal filters.
Deep eutectic solvent formulation
The deep eutectic solvents 1:2 choline chloride:glycerol, 1:4 tetrabutylammonium chloride:glycerol, and 1:2 tetrabutylammonium chloride:glycerol and the deuterated analogs were prepared by mixing the components and heating at 60 °C under an argon atmosphere until a homogeneous liquid was formed. Data for the deuterated molecules that support the findings of this study are openly available via 10.5281/zenodo.14202086. The residual water content in the neat DESs was determined using Karl−Fischer titration and found to be 0.42 ± 0.06 wt%, 0.28 ± 0.12 wt%, and 0.22 ± 0.08 wt% water for ChCl:Glyc, TBAC:Glyc, and TBAC:OA respectively.
Small-angle neutron scattering
SANS experiments were performed on D33 at Institut Laue-Langevin—The European Neutron Source. Data of protiated [C-Mb][S] in deuterated solvents was collected in the q-range between 0.005 and 0.33 Å-1. Data reduced with Grasp51, and were analyzed using SasView 5.0. The experiment was conducted under the awarded beamtime 9-13-106252.
Spectroscopy methods
UV−vis spectroscopy was conducted using a Shimazdu UV-2600i Spectrophotometer for protein characterization and a Implen NanoPhotometer N60 Spectrophotometer for the determination of the Kamlet-Taft parameters. Data was collected between 220 nm and 800 nm.
In house CD measurements were performed on a Chirascan V100 spectrometer in the wavelength range between 180 and 280 nm. SRCD measurements were performed on the beamline B23 at Diamond Light Source using a custom Linkam stage for temperature control under the awarded beamtime SM33225. Data was acquired in between 180 nm and 260 nm. CD spectra were analyzed using the platform BeStSel37.
Samples were loaded in quartz cuvettes and the pathlength was selected to optimize the signal-to-noise ratio for each technique.
Further details on the experimental aspects and data analyses are presented in the Supplementary Methods.
Data availability
Data (ASCII files) are openly available under the Digital Object Identifier (DOI): https://doi.org/10.5281/zenodo.10548643.
References
Yadav, N. & Venkatesu, P. Current understanding and insights towards protein stabilization and activation in deep eutectic solvents as sustainable solvent media. Phys. Chem. Chem. Phys. 24, 13474–13509 (2022).
Martins, M. A. R., Pinho, S. P. & Coutinho, J. A. P. Insights into the nature of eutectic and deep eutectic mixtures. J. Solut. Chem. 48, 962–982 (2019).
Hansen, B. B. et al. Deep eutectic solvents: a review of fundamentals and applications. Chem. Rev. 121, 1232–1285 (2021).
Pandey, A., Rai, R., Pal, M. & Pandey, S. How polar are choline chloride-based deep eutectic solvents? Phys. Chem. Chem. Phys. 16, 1559–1568 (2014).
Zahn, S., Kirchner, B. & Mollenhauer, D. Charge spreading in deep eutectic solvents. Chemphyschem 17, 3354–3358 (2016).
Dwamena, A. K. & Raynie, D. E. Solvatochromic parameters of deep eutectic solvents: effect of different carboxylic acids as hydrogen bond donor. J. Chem. Eng. Data 65, 640–646 (2020).
Esquembre, R. et al. Thermal unfolding and refolding of lysozyme in deep eutectic solvents and their aqueous dilutions. Phys. Chem. Chem. Phys. 15, 11248–11256 (2013).
Sanchez-Fernandez, A., Edler, K. J., Arnold, T., Alba Venero, D. & Jackson, A. J. Protein conformation in pure and hydrated deep eutectic solvents. Phys. Chem. Chem. Phys. 19, 8667–8670 (2017).
Gorke, J. T., Srienc, F. & Kazlauskas, R. J. Hydrolase-catalyzed biotransformations in deep eutectic solvents. Chem Commun (Camb), 1235–1237 (2008).
Toledo, M. L. et al. Laccase activation in deep eutectic solvents. ACS Sustain. Chem. Eng. 7, 11806–11814 (2019).
Sanchez-Fernandez, A., Prevost, S. & Wahlgren, M. Deep eutectic solvents for the preservation of concentrated proteins: the case of lysozyme in 1 : 2 choline chloride : glycerol. Green. Chem. 24, 4437–4442 (2022).
Dhiman, D. et al. Unveiling the potential of deep eutectic solvents to improve the conformational and colloidal stability of immunoglobulin G antibodies. Green. Chem. 25, 650–660 (2023).
Belviso, B. D. et al. Introducing protein crystallization in hydrated deep eutectic solvents. ACS Sustain. Chem. Eng. 9, 8435–8449 (2021).
Zhang, Y., Wang, Y., Guan, Y. & Zhang, Y. Peptide-enhanced tough, resilient and adhesive eutectogels for highly reliable strain/pressure sensing under extreme conditions. Nat. Commun. 13, 6671 (2022).
Picchio, M. L. et al. Elastomeric protein bioactive eutectogels for topical drug delivery. Adv. Funct. Mater., 2313747 (2024).
Sanchez-Fernandez, A., Poon, J.-F., Leung, A. E., Prévost, S. F. & Dicko, C. Stabilization of non-native folds and programmable protein gelation in compositionally designed deep eutectic solvents. ACS Nano 18, 18314–18326 (2024).
Sanchez-Fernandez, A. et al. Hydration in deep eutectic solvents induces non-monotonic changes in the conformation and stability of proteins. J. Am. Chem. Soc. 144, 23657–23667 (2022).
Kumari, M., Kumari, P. & Kashyap, H. K. Structural adaptations in the bovine serum albumin protein in archetypal deep eutectic solvent reline and its aqueous mixtures. Phys. Chem. Chem. Phys. 24, 5627–5637 (2022).
Zhang, L. et al. Kamlet–Taft parameters of deep eutectic solvents and their relationship with dissolution of main lignocellulosic components. Ind. Eng. Chem. Res. 62, 11723–11734 (2023).
Aroso, I. M. et al. Dissolution enhancement of active pharmaceutical ingredients by therapeutic deep eutectic systems. Eur. J. Pharm. Biopharm. 98, 57–66 (2016).
Guajardo, N., Müller, C. R., Schrebler, R., Carlesi, C. & Domínguez de María, P. Deep eutectic solvents for organocatalysis, biotransformations, and multistep organocatalyst/enzyme combinations. ChemCatChem 8, 1020–1027 (2015).
Florindo, C., Branco, L. C. & Marrucho, I. M. Quest for green-solvent design: from hydrophilic to hydrophobic (deep) eutectic solvents. ChemSusChem 12, 1549–1559 (2019).
Brogan, A. P. S., Bui-Le, L. & Hallett, J. P. Non-aqueous homogenous biocatalytic conversion of polysaccharides in ionic liquids using chemically modified glucosidase. Nat. Chem. 10, 859–865 (2018).
Anselmo, A. C., Gokarn, Y. & Mitragotri, S. Non-invasive delivery strategies for biologics. Nat. Rev. Drug Discov. 18, 19–40 (2019).
Wang, X. et al. Corner engineering: tailoring enzymes for enhanced resistance and thermostability in deep eutectic solvents. Angew. Chem. Int. Ed. Engl. 63, e202315125 (2024).
Sheng, Y. et al. Harnessing solvation-guided engineering to enhance deep eutectic solvent resistance and thermostability in enzymes. Green. Chem. 26, 9132–9141 (2024).
Brogan, A. P. S., Siligardi, G., Hussain, R., Perriman, A. W. & Mann, S. Hyper-thermal stability and unprecedented re-folding of solvent-free liquid myoglobin. Chem. Sci. 3, 1839–1846 (2012).
Brogan, A. P. & Hallett, J. P. Solubilizing and stabilizing proteins in anhydrous ionic liquids through formation of protein-polymer surfactant nanoconstructs. J. Am. Chem. Soc. 138, 4494–4501 (2016).
Brogan, A. P. S. Preparation and application of solvent-free liquid proteins with enhanced thermal and anhydrous stabilities. N. J. Chem. 45, 6577–6585 (2021).
Meza Huaman, S. M., Nicholson, J. H. & Brogan, A. P. S. A general route to retooling hydrolytic enzymes toward plastic degradation. Cell Rep. Phys. Sci. 5 (2024).
Peterson, E. S. et al. Folding myoglobin within a sol-gel glass: protein folding constrained to a small volume. Biophys. J. 95, 322–332 (2008).
Cort, J. R. & Andersen, N. H. Formation of a molten-globule-like state of myoglobin in aqueous hexafluoroisopropanol. Biochem. Biophys. Res. Commun. 233, 687–691 (1997).
Andersen, K. K., Westh, P. & Otzen, D. E. Global study of myoglobin−surfactant interactions. Langmuir 24, 399–407 (2008).
Abbott, A. P. et al. Glycerol eutectics as sustainable solvent systems. Green. Chem. 13, 82–90 (2011).
Mjalli, F. S., Naser, J., Jibril, B., Alizadeh, V. & Gano, Z. Tetrabutylammonium chloride based ionic liquid analogues and their physical properties. J. Chem. Eng. Data 59, 2242–2251 (2014).
Teles, A. R. R. et al. Solvatochromic parameters of deep eutectic solvents formed by ammonium-based salts and carboxylic acids. Fluid Phase Equilib. 448, 15–21 (2017).
Micsonai, A. et al. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. USA 112, E3095–E3103 (2015).
Sanchez-Fernandez, A., Jackson, A. J., Prevost, S. F., Doutch, J. J. & Edler, K. J. Long-range electrostatic colloidal interactions and specific ion effects in deep eutectic solvents. J. Am. Chem. Soc. 143, 14158–14168 (2021).
Sanchez-Fernandez, A. et al. An integrative toolbox to unlock the structure and dynamics of protein-surfactant complexes. Nanoscale Adv. 2, 4011–4023 (2020).
Sinden, R. R. in DNA Structure and Function (ed Richard R. Sinden) 287-325 (Academic Press, 1994).
Havlin, S. & Ben-Avraham, D. Fractal dimensionality of polymer chains. J. Phys. A: Math. Gen. 15, L311 (1982).
Javadpour, M. M. et al. De novo antimicrobial peptides with low mammalian cell toxicity. J. Med. Chem. 39, 3107–3113 (1996).
Farooq, M. Q., Abbasi, N. M. & Anderson, J. L. Deep eutectic solvents in separations: Methods of preparation, polarity, and applications in extractions and capillary electrochromatography. J. Chromatogr. A 1633, 461613 (2020).
Florindo, C., McIntosh, A. J. S., Welton, T., Branco, L. C. & Marrucho, I. M. A closer look into deep eutectic solvents: exploring intermolecular interactions using solvatochromic probes. Phys. Chem. Chem. Phys. 20, 206–213 (2017).
Sanchez-Fernandez, A. et al. Surfactant-solvent interaction effects on the micellization of cationic surfactants in a carboxylic acid-based deep eutectic solvent. Langmuir 33, 14304–14314 (2017).
Sanchez-Fernandez, A. et al. Micelle structure in a deep eutectic solvent: a small-angle scattering study. Phys. Chem. Chem. Phys. 18, 14063–14073 (2016).
Manasi, I., King, S. M. & Edler, K. J. Cationic micelles in deep eutectic solvents: effects of solvent composition. Faraday Discuss 253, 26–41 (2024).
Sanchez-Fernandez, A., Leung, A. E., Kelley, E. G. & Jackson, A. J. Complex by design: hydrotrope-induced micellar growth in deep eutectic solvents. J. Colloid Interface Sci. 581, 292–298 (2021).
Atri, R. S. et al. Morphology modulation of ionic surfactant micelles in ternary deep eutectic solvents. J. Phys. Chem. B 124, 6004–6014 (2020).
Stefanovic, R., Webber, G. B. & Page, A. J. Polymer solvation in choline chloride deep eutectic solvents modulated by the hydrogen bond donor. J. Mol. Liq. 279, 584–593 (2019).
Dewhurst, C. D. Graphical reduction and analysis small-angle neutron scattering program: GRASP. J. Appl. Crystallogr. 56, 1595–1609 (2023).
Sanchez-Fernandez, A., Brogan, A. P. S., Meza Huaman, S. M., Nicholson, J. & Prevost, S. Solubilising protein nanoconstructs in deep eutectic solvents with tailored hydrophobicity. Institut Laue-Langevin (ILL), 2023, https://doi.org/10.5291/ILL-DATA.9-13-1062.
Acknowledgements
We are thankful for the financial support of Spanish grants RYC2022-037909-I and PID2022-141673OA-I00 funded by MCIN/AEI and by “ERDF A way of making Europe”. This project has received funding from the European Union’s Horizon research and innovation programme under the Marie Skłodowska-Curie grant agreement No 101063372, and was supported by funds provided by King’s College London. The authors thank the Institute Laue-Langevin Langevin—The European Neutron Source (https://doi.org/10.5291/ILL-DATA.9-13-1062) and Diamond Light Source (SM33225) for the awarded beamtime and access to D33 and B23 respectively.
Author information
Authors and Affiliations
Contributions
A.S.-F. and A.P.S.B. conceived the idea and designed the experiments. A.S.-F., J.H.N., S.M.M.H., and C.A.R. performed the experiments and analyzed the data. J.-F.P. prepared the deuterated compounds used during the SANS experiments. S.P. assisted during the SANS experiments. A.S.-F. and A.P.S.B. wrote the manuscript. All authors proof-read, discussed and have given approval to the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Chemistry thanks Iva Manasi and the other, anonymous, reviewers for their contribution to the peer review of this work. Peer review reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sanchez-Fernandez, A., Nicholson, J.H., Meza Huaman, S.M. et al. Unlocking the full compositional control of hydrophilic and hydrophobic deep eutectic solvents over protein structure and stability. Commun Chem 8, 173 (2025). https://doi.org/10.1038/s42004-025-01571-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42004-025-01571-6