Abstract
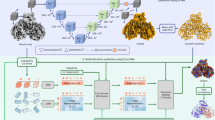
While cryogenic-electron microscopy yields high-resolution density maps for complex structures, accurate determination of the corresponding atomic structures still necessitates significant expertise and labour-intensive manual interpretation. Recently, artificial intelligence-based methods have emerged to streamline this process; however, several challenges persist. First, existing methods typically require multi-stage training and inference, causing inefficiencies and inconsistency. Second, these approaches often encounter bias and incur substantial computational costs in aligning predicted atomic coordinates with sequence. Last, due to the limitations of available datasets, previous studies struggle to generalize effectively to complicated and unseen test data. Here, in response to these challenges, we introduce end-to-end and efficient CryoFold (E3-CryoFold), a deep learning method that enables end-to-end training and one-shot inference. E3-CryoFold uses three-dimensional and sequence transformers to extract features from density maps and sequences, using cross-attention modules to integrate the two modalities. Additionally, it uses an SE(3) graph neural network to construct atomic structures based on extracted features. E3-CryoFold incorporates a pretraining stage, during which models are trained on simulated density maps derived from Protein Data Bank structures. Empirical results demonstrate that E3-CryoFold improves the average template modelling score of the generated structures by 400% as compared to Cryo2Struct and significantly outperforms ModelAngelo, while achieving this huge improvement using merely one-thousandth of the inference time required by these methods. Thus, E3-CryoFold represents a robust, streamlined and cohesive framework for cryogenic-electron microscopy structure determination.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The experimental dataset can be downloaded at https://doi.org/10.7910/DVN/FCDG0W (ref. 46), and the standard test dataset can be downloaded at https://doi.org/10.7910/DVN/2GSSC9 (ref. 47). The low-resolution and simulated datasets are accessible at https://zhanggroup.org/CR-I-TASSER/. All source data are accessible from ref. 48 (standard_test_data.xlsx for standard test dataset, novel_test_data.xlsx for novel established test dataset, low_resolution_experimental_data.xlsx for low-resolution density maps, simulated_data.xlsx for simulated density maps). Source data are provided with this paper.
Code availability
The source code of E3-CryoFold is available via GitHub at https://github.com/A4Bio/CryoFold/ (ref. 49). This repository also contains the instructions and tutorial for applying E3-CryoFold on an example cryo-EM map to generate a complex structure.
Change history
22 August 2025
In the version of the article initially published, in the Acknowledgements, the National Science and Technology Major Project grant number was incorrect and has now been amended to 2022ZD0115102 in the HTML and PDF versions of the article.
11 July 2025
A Correction to this paper has been published: https://doi.org/10.1038/s42256-025-01094-8
References
Boadu, F., Cao, H. & Cheng, J. Combining protein sequences and structures with transformers and equivariant graph neural networks to predict protein function. Bioinformatics 39, i318–i325 (2023).
Bai, X.-C., McMullan, G. & Scheres, S. H. How cryo-EM is revolutionizing structural biology. Trends Biochem. Sci. 40, 49–57 (2015).
Lawson, C. L. et al. Outcomes of the EMDataResource cryo-EM ligand modeling challenge. Nat. Methods 21, 1340–1348 (2024).
Dhakal, A., Gyawali, R., Wang, L. & Cheng, J. A large expert-curated cryo-EM image dataset for machine learning protein particle picking. Sci. Data 10, 392 (2023).
Dhakal, A., Gyawali, R., Wang, L. & Cheng, J. Cryotransformer: a transformer model for picking protein particles from cryo-EM micrographs. Bioinformatics 40, btae109 (2024).
Giri, N., Roy, R. S. & Cheng, J. Deep learning for reconstructing protein structures from cryo-EM density maps: recent advances and future directions. Curr. Opin. Struct. Biol. 79, 102536 (2023).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 486–501 (2010).
Croll, T. I. Isolde: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. Sect. D: Struct. Biol. 74, 519–530 (2018).
Gao, Y., Thorn, V. & Thorn, A. Errors in structural biology are not the exception. Acta Crystallogr. Sect. D Struct. Biol. 79, 206–211 (2023).
Croll, T. I. et al. Making the invisible enemy visible. Nat. Struct. Mol. Biol. 28, 404–408 (2021).
Zhong, E. D., Bepler, T., Berger, B. & Davis, J. H. Cryodrgn: reconstruction of heterogeneous cryo-EM structures using neural networks. Nat. Methods 18, 176–185 (2021).
Rangan, R. et al. CryoDRGN-ET: deep reconstructing generative networks for visualizing dynamic biomolecules inside cells. Nat. Methods 21, 1537–1545 (2024).
Levy, A., Wetzstein, G., Martel, J. N., Poitevin, F. & Zhong, E. Amortized inference for heterogeneous reconstruction in cryo-EM. Adv. Neural Inf. Process. Syst. 35, 13038–13049 (2022).
Pfab, J., Phan, N. M. & Si, D. Deeptracer for fast de novo cryo-EM protein structure modeling and special studies on cov-related complexes. Proc. Natl Acad. Sci. USA 118, e2017525118 (2021).
Ronneberger, O., Fischer, P. & Brox, T. U-net: convolutional networks for biomedical image segmentation. In Proc. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Part III Vol. 18, 234–241 (Springer, 2015).
Hoffman, K. L. & Padberg, M. et al. Traveling salesman problem. Encycl. Oper. Res. Manag. Sci. 1, 1573–1578 (2013).
Jamali, K. et al. Automated model building and protein identification in cryo-EM maps. Nature 628, 450–457 (2024).
Rabiner, L. & Juang, B. An introduction to hidden Markov models. IEEE ASSP Mag. 3, 4–16 (1986).
Terashi, G., Wang, X., Prasad, D., Nakamura, T. & Kihara, D. Deepmainmast: integrated protocol of protein structure modeling for cryo-EM with deep learning and structure prediction. Nat. Methods 21, 122–131 (2024).
Jumper, J. et al. Highly accurate protein structure prediction with alphafold. Nature 596, 583–589 (2021).
Giri, N. & Cheng, J. De novo atomic protein structure modeling for cryoEM density maps using 3D transformer and HMM. Nat. Commun. 15, 5511 (2024).
Vaswani, A. Attention is all you need. In Proc. Advances in Neural Information Processing Systems Vol. 30 (eds Guyon, I. et al.) 6000–6010 (Curran Associates, 2017).
Shekhar, M. et al. Cryofold: determining protein structures and data-guided ensembles from cryo-em density maps. Matter 4, 3195–3216 (2021).
Lawson, C. L. et al. Emdatabank unified data resource for 3DEM. Nucleic Acids Res. 44, D396–D403 (2016).
Zhang, Y. & Skolnick, J. Tm-align: a protein structure alignment algorithm based on the Tm-score. Nucleic Acids Res. 33, 2302–2309 (2005).
Chen, C.-F. R., Fan, Q. & Panda, R. Crossvit: cross-attention multi-scale vision transformer for image classification. In Proc. IEEE/CVF International Conference on Computer Vision 357–366 (IEEE, 2021).
Satorras, V. G., Hoogeboom, E. & Welling, M. E (n) equivariant graph neural networks. In Proc. International Conference on Machine Learning 9323–9332 (PMLR, 2021).
Han, J., Rong, Y., Xu, T. & Huang, W. Geometrically equivariant graph neural networks: a survey. Preprint at https://arxiv.org/abs/2202.07230 (2022).
Giri, N., Wang, L. & Cheng, J. Cryo2StructData: a large labeled cryo-EM density map dataset for AI-based modeling of protein structures. Sci. Data 11, 458 (2024).
Terwilliger, T. C., Adams, P. D., Afonine, P. V. & Sobolev, O. V. A fully automatic method yielding initial models from high-resolution cryo-electron microscopy maps. Nat. Methods 15, 905–908 (2018).
Mariani, V., Biasini, M., Barbato, A. & Schwede, T. LDDT: a local superposition-free score for comparing protein structures and models using distance difference tests. Bioinformatics 29, 2722–2728 (2013).
Basu, S. & Wallner, B. DockQ: a quality measure for protein-protein docking models. PLoS ONE 11, e0161879 (2016).
Pintilie, G. et al. Measurement of atom resolvability in cryo-EM maps with Q-scores. Nat. Methods 17, 328–334 (2020).
Van Heel, M. & Schatz, M. Fourier shell correlation threshold criteria. J. Struct. Biol. 151, 250–262 (2005).
Yamashita, K., Palmer, C. M., Burnley, T. & Murshudov, G. N. Cryo-EM single-particle structure refinement and map calculation using servalcat. Biol. Crystallogr. 77, 1282–1291 (2021).
Allegretti, M., Mills, D. J., McMullan, G., Kühlbrandt, W. & Vonck, J. Atomic model of the F420-reducing [NiFe] hydrogenase by electron cryo-microscopy using a direct electron detector. eLife 3, e01963 (2014).
Bartesaghi, A., Matthies, D., Banerjee, S., Merk, A. & Subramaniam, S. Structure of β-galactosidase at 3.2-Å resolution obtained by cryo-electron microscopy. Proc. Natl Acad. Sci. USA 111, 11709–11714 (2014).
Hattne, J. et al. Analysis of global and site-specific radiation damage in cryo-EM. Structure 26, 759–766 (2018).
Lin, Z. et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 379, 1123–1130 (2023).
Young, I. T. & Van Vliet, L. J. Recursive implementation of the Gaussian filter. Signal Process. 44, 139–151 (1995).
Loshchilov, I. et al. Fixing weight decay regularization in Adam. Preprint at https://arxiv.org/abs/1711.05101 (2017).
Smith, L. N. Cyclical learning rates for training neural networks. In Proc. 2017 IEEE Winter Conference on Applications of Computer Vision (WACV) 464–472 (IEEE, 2017).
Gao, Z., Tan, C. & Li, S. Z. Foldtoken4: Consistent & hierarchical fold language. Preprint at bioRxiv https://doi.org/10.1101/2024.08.04.606514 (2024).
Ingraham, J. B. et al. Illuminating protein space with a programmable generative model. Nature 623, 1070–1078 (2023).
Chomboon, K., Chujai, P., Teerarassamee, P., Kerdprasop, K. & Kerdprasop, N. An empirical study of distance metrics for k-nearest neighbour algorithm. In Proc. 3rd International Conference on Industrial Application Engineering Vol. 2, p. 4 (Institute of Industrial Applications Engineers, 2015).
Giri, N., Wang, L. & Cheng, J. Cryo2StructData: full dataset. Harvard Dataverse https://doi.org/10.7910/DVN/FCDG0W (2023).
Giri, N., Wang, L. & Cheng, J. Cryo2StructData: test dataset. Harvard Dataverse https://doi.org/10.7910/DVN/2GSSC9 (2023).
Wang, J. cryofold_source_data.zip. figshare https://doi.org/10.6084/m9.figshare.28530359.v1 (2025).
Wang, J. & Tan, C. End-to-end Cryo-EM complex structure determination with high accuracy and ultra-fast speed. Zenodo https://doi.org/10.5281/zenodo.14970359 (2025).
Hodson, T. O. Root mean square error (RMSE) or mean absolute error (MAE): when to use them or not. Geosci. Model Dev. 15, 5481–5487 (2022).
Acknowledgements
This work was supported by National Science and Technology Major Project (grant no. 2022ZD0115102), National Natural Science Foundation of China Project (grant no. 624B2115, and grant no. U21A20427), Project (grant no. WU2022A009) from the Center of Synthetic Biology and Integrated Bioengineering of Westlake University and Project (grant no. WU2023C019) from the Westlake University Industries of the Future Research Funding.
Author information
Authors and Affiliations
Contributions
J.W. conceived the idea and developed the framework. Z.G. provided the crucial technology for SE(3) GNN and structure generation, as well as the pretraining dataset. J.W. drafted the paper and C.T. helped in writing Methods. C.T., Z.G., Y.Z., G.Z. helped in editing the paper. J.W. and C.T. prepared codes and released them on GitHub. S.Z.L. supervised the project and helped revise the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Machine Intelligence thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary information for E3-CryoFold.
Supplementary Data 1
Statistical data for Supplementary Fig. 2.
Supplementary Data 2
Statistical data for Supplementary Fig. 3.
Source data
Source Data Fig. 2
Statistical source data for Fig. 2.
Source Data Fig. 4
Statistical source data for Fig. 4.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, J., Tan, C., Gao, Z. et al. End-to-end cryo-EM complex structure determination with high accuracy and ultra-fast speed. Nat Mach Intell 7, 1091–1103 (2025). https://doi.org/10.1038/s42256-025-01056-0
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s42256-025-01056-0