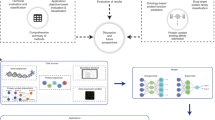
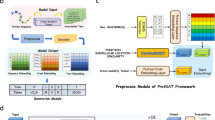
Abstract
Protein language models are trained to predict amino acid sequences from vast protein databases and learn to represent proteins as feature vectors. These vector representations have enabled impressive applications, from predicting mutation effects to protein folding. One of the reasons offered for the success of these models is that conserved sequence motifs tend to be important for protein fitness. Yet, the relationship between sequence conservation and fitness can be confounded by the evolutionary and environmental context. Should we, therefore, look to other data sources that may contain more direct functional information? In this work, we conduct a comprehensive study examining the effects of training protein models to predict 19 types of text annotation from UniProt. Our results show that fine-tuning protein models on a subset of these annotations enhances the models’ predictive capabilities on a variety of function prediction tasks. In particular, when evaluated on our tasks, our model outperforms the basic local alignment search tool, which none of the pretrained protein models accomplished. Our results suggest that a much wider array of data modalities, such as text annotations, may be tapped to improve protein language models.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The raw Swiss-Prot dataset can be downloaded from UniProt (https://ftp.uniprot.org/pub/databases/uniprot/previous_major_releases/release-2023_02/knowledgebase/). Our parsed and processed version used for pretraining is available at https://huggingface.co/datasets/mskrt/PAIR/. The figshare repository is available at https://doi.org/10.6084/m9.figshare.27004768. Binary localization, Subcellular location and Fold datasets used in the ‘PAIR improves protein function predictions’ section can be downloaded through the TorchDrug library (https://github.com/DeepGraphLearning/torchdrug). The DTI datasets DAVIS and BindingDB are available from Therapeutics Data Commons (https://tdcommons.ai/overview). Source data are provided with this paper.
Code availability
The code is publicly available via GitHub at https://github.com/h4duan/PAIR. A preserved version of the code is available via Zenodo at https://doi.org/10.5281/zenodo.14834853 (ref. 44). The model checkpoints are publicly available at https://huggingface.co/h4duan.
References
Consortium, T. U. UniProt: the universal protein knowledgebase in 2025. Nucleic Acids Res. 53, D609–D617 (2025).
Rives, A. et al. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. Proc. Natl Acad. Sci. USA 118, e2016239118 (2021).
Lin, Z. et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 379, 1123–1130 (2023).
Elnaggar, A. et al. ProtTrans: toward understanding the language of life through self-supervised learning. IEEE Trans. Pattern Anal. Mach. Intell. 44, 7112–7127 (2021).
Yu, T. et al. Enzyme function prediction using contrastive learning. Science 379, 1358–1363 (2023).
Hie, B. L. et al. Efficient evolution of human antibodies from general protein language models. Nat. Biotechnol. 42, 275–283 (2024).
Hopf, T. A. et al. Mutation effects predicted from sequence co-variation. Nat. Biotechnol. 35, 128–135 (2017).
Wei, J. et al. Emergent abilities of large language models. Trans. Mach. Learn. Res. 2022, 1–30 (2022).
Maddison, W. P. & FitzJohn, R. G. The unsolved challenge to phylogenetic correlation tests for categorical characters. Syst. Biol. 64, 127–136 (2014).
Vu, M. H. et al. Linguistically inspired roadmap for building biologically reliable protein language models. Nat. Mach. Intell. 5, 485–496 (2023).
Xu, M., Yuan, X., Miret, S. & Tang, J. ProtST: multi-modality learning of protein sequences and biomedical texts. In International Conference on Machine Learning 38749–38767 (PMLR, 2023).
Liu, S. et al. A text-guided protein design framework. Nat. Mach. Intell. 7, 580–591 (2025).
Zhang, N. et al. OntoProtein: protein pretraining with Gene Ontology embedding. In International Conference on Learning Representations 1–18 (2022).
You, R., Huang, X. & Zhu, S. DeepText2GO: improving large-scale protein function prediction with deep semantic text representation. Methods 145, 82–90 (2018).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Hamamsy, T. et al. Protein remote homology detection and structural alignment using deep learning. Nat. Biotechnol. 42, 975–985 (2024).
Rothe, S., Narayan, S. & Severyn, A. Leveraging pre-trained checkpoints for sequence generation tasks. Trans. Assoc. Comput. Linguist. 8, 264–280 (2020).
Beltagy, I., Lo, K. & Cohan, A. SciBERT: a pretrained language model for scientific text. In Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP) 3615–3620 (Association for Computational Linguistics, 2019).
Luong, M., Le, Q. V., Sutskever, I., Vinyals, O. & Kaiser, L. Multi-task sequence to sequence learning. In 4th International Conference on Learning Representations 1–10 (ICLR, 2016).
Almagro Armenteros, J. J., Sønderby, C. K., Sønderby, S. K., Nielsen, H. & Winther, O. DeepLoc: prediction of protein subcellular localization using deep learning. Bioinformatics 33, 3387–3395 (2017).
Hou, J., Adhikari, B. & Cheng, J. DeepSF: deep convolutional neural network for mapping protein sequences to folds. Bioinformatics 34, 1295–1303 (2018).
Davis, M. I. et al. Comprehensive analysis of kinase inhibitor selectivity. Nat. Biotechnol. 29, 1046–1051 (2011).
Liu, T., Lin, Y., Wen, X., Jorissen, R. N. & Gilson, M. K. BindingDB: a web-accessible database of experimentally determined protein–ligand binding affinities. Nucleic Acids Res. 35, D198–D201 (2007).
Xu, M. et al. PEER: a comprehensive and multi-task benchmark for protein sequence understanding. Adv. Neural Inf. Process. Syst. 35, 35156–35173 (2022).
Christofidellis, D. et al. Unifying molecular and textual representations via multi-task language modelling. In Proc. 40th International Conference on Machine Learning 202, 6140–6157 (International Conference on Machine Learning, 2023).
Vaswani, A. et al. Attention is all you need. Adv. Neural Inf. Process. Syst. 30, 5998–6008 (2017).
Wolf, T. et al. Transformers: state-of-the-art natural language processing. In Proc. 2020 Conference on Empirical Methods in Natural Language Processing: System Demonstrations 38–45 (Association for Computational Linguistics, 2020).
Dai, Z. et al. Transformer-XL: attentive language models beyond a fixed-length context. In Proc. 57th Annual Meeting of the Association for Computational Linguistics 2978–2988 (Association for Computational Linguistics, 2019).
Suzek, B. E. et al. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 31, 926–932 (2015).
Steinegger, M. & Söding, J. Clustering huge protein sequence sets in linear time. Nat. Commun. 9, 2542 (2018).
Devlin, J., Chang, M.-W., Lee, K. & Toutanova, K. BERT: pre-training of deep bidirectional transformers for language understanding. In Proc. 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies 4171–4186 (Association for Computational Linguistics, 2019).
Kalamkar, D. et al. A study of BFLOAT16 for deep learning training. Preprint at https://arxiv.org/abs/1905.12322 (2019).
Loshchilov, I. & Hutter, F. Decoupled weight decay regularization. In International Conference on Learning Representations 1–8 (2018).
Kulmanov, M. & Hoehndorf, R. DeepGOZero: improving protein function prediction from sequence and zero-shot learning based on ontology axioms. Bioinformatics 38, i238–i245 (2022).
Gane, A. et al. ProtNLM: model-based natural language protein annotation (2023); https://www.uniprot.org/help/ProtNLM
Huang, K. et al. Therapeutics data commons: machine learning datasets and tasks for drug discovery and development. In Thirty-Fifth Conference on Neural Information Processing Systems Datasets and Benchmarks Track (Round 1) 1–22 (NeurIPS, 2021).
You, R. et al. GOLabeler: improving sequence-based large-scale protein function prediction by learning to rank. Bioinformatics 34, 2465–2473 (2018).
BLAST+ executable (2024); https://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/
Sanderson, T., Bileschi, M. L., Belanger, D. & Colwell, L. J. ProteInfer, deep neural networks for protein functional inference. eLife 12, e80942 (2023).
Taylor, R. et al. Galactica: a large language model for science. Preprint at https://arxiv.org/abs/2211.09085 (2022).
Microsoft Research AI4Science & Microsoft Azure Quantum. The impact of large language models on scientific discovery: a preliminary study using GPT-4. Preprint at https://arxiv.org/abs/2311.07361 (2023).
M. Bran, A. et al. Augmenting large language models with chemistry tools. Nat. Mach. Intell. 6, 525–535 (2024).
Mirza, A. et al. Are large language models superhuman chemists? Preprint at https://arxiv.org/abs/2404.01475 (2024).
Duan, H. et al. Boosting the predictive power of protein representations with a corpus of text annotations. Zenodo https://doi.org/10.5281/zenodo.14834853 (2025).
Karamcheti, S. et al. Prismatic VLMs: investigating the design space of visually-conditioned language models. In Proc. 41st International Conference on Machine Learning 930 (JMLR, 2024).
Acknowledgements
We would like to thank C. Harrigan, A. Jung and Y. Ruan for insightful discussions. This work was supported in part by Advanced Micro Devices, Inc. under the AMD AI&HPC Fund program, as well as by the Acceleration Consortium and the Vector Institute. A.A.G. thanks A. G. Frøseth for his generous support, as well as Natural Resources Canada and the Canada 150 Research Chairs program (NSERC-IRCPJ 547644). The research was enabled in part by the computational resources provided by the Vector Institute for Artificial Intelligence (https://vectorinstitute.ai/) and the Acceleration Consortium (https://acceleration.utoronto.ca/).
Author information
Authors and Affiliations
Contributions
H.D. led the model aspect of the project. M.S. led the data component. H.D. and M.S. co-led the evaluation part. L.C. designed the data curation pipeline and provided advisory input across multiple aspects of the project. E.M.R. contributed to the data curation, biological evaluation and conception of this work. N.D. contributed to the model implementation and implemented the baseline for evaluation. A.A.-G. contributed to the conception of the work and provided supervision. C.J.M. served as the main supervisor, contributed to the blueprint of the work and advised on all pipelines. All authors contributed to the writing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Machine Intelligence thanks Ankur Parikh and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Sections A–E, Figs. A and B and Tables A–C.
Source data
Source Data Figs. 2–5
Raw experimental data presented in Figs. 2–5. Each worksheet is named according to the specific figure it corresponds to.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Duan, H., Skreta, M., Cotta, L. et al. Boosting the predictive power of protein representations with a corpus of text annotations. Nat Mach Intell 7, 1403–1413 (2025). https://doi.org/10.1038/s42256-025-01088-6
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s42256-025-01088-6
This article is cited by
-
Protein foundation models: a comprehensive survey
Science China Life Sciences (2026)