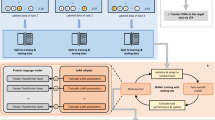
Abstract
Protein engineering holds substantial promise for designing proteins with customized functions, yet the vast landscape of potential mutations versus limited laboratory capacity constrains the discovery of optimal sequences. Here, to address this, we present the μProtein framework, which accelerates protein engineering by combining μFormer, a deep learning model for accurate mutational effect prediction, with μSearch, a reinforcement learning algorithm designed to efficiently navigate the protein fitness landscape using μFormer as an oracle. μProtein leverages single-mutation data to predict optimal sequences with complex, multi-amino-acid mutations through its modelling of epistatic interactions and a multi-step search strategy. In addition to strong performance on benchmark datasets, μProtein identified high-gain-of-function multi-point mutants for the enzyme β-lactamase, surpassing one of the highest-known activity levels, in wet laboratory, trained solely on single-mutation data. These results demonstrate μProtein’s capability to discover impactful mutations across the vast protein sequence space, offering a robust, efficient approach for protein optimization.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The FLIP benchmark is publicly available at http://data.bioembeddings.com/public/FLIP/. The ProteinGym v0.1 data collection can be accessed via Hugging Face at https://huggingface.co/datasets/OATML-Markslab/ProteinGym_v0.1. The dataset under our split scheme is available via figshare at https://doi.org/10.6084/m9.figshare.26892355 (ref. 69). The list of ESBLs are available at http://bldb.eu/ (ref. 44). The fitness scores for β-lactamase variants against cefotaxime, validated by our wet-laboratory experiments, are provided in the Supplementary Information. Source data are provided with this paper.
Code availability
The code to reproduce the results of this paper is publicly available via GitHub at https://github.com/microsoft/Mu-Protein and via Zenodo at https://doi.org/10.5281/zenodo.15836168 (ref. 76). The data-split scheme is available via figshare at https://doi.org/10.6084/m9.figshare.26892355 (ref. 69).
References
Miton, C. M. & Tokuriki, N. Insertions and deletions (indels): a missing piece of the protein engineering jigsaw. Biochemistry 62, 148–157 (2023).
Gray, V. E., Hause, R. J., Luebeck, J., Shendure, J. & Fowler, D. M. Quantitative missense variant effect prediction using large-scale mutagenesis data. Cell Syst. 6, 116–124 (2018).
Riesselman, A. J., Ingraham, J. B. & Marks, D. S. Deep generative models of genetic variation capture the effects of mutations. Nat. Methods 15, 816–822 (2018).
Biswas, S., Khimulya, G., Alley, E. C., Esvelt, K. M. & Church, G. M. Low-n protein engineering with data-efficient deep learning. Nat. Methods 18, 389–396 (2021).
Notin, P. et al. Tranception: protein fitness prediction with autoregressive transformers and inference-time retrieval. In Proc. 39th International Conference on Machine Learning 16990–17017 (PMLR, 2022).
Fowler, D. M. & Fields, S. Deep mutational scanning: a new style of protein science. Nat. Methods 11, 801–807 (2014).
Weile, J. & Roth, F. P. Multiplexed assays of variant effects contribute to a growing genotype–phenotype atlas. Hum. Genet. 137, 665–678 (2018).
Wittmund, M., Cadet, F. & Davari, M. D. Learning epistasis and residue coevolution patterns: current trends and future perspectives for advancing enzyme engineering. ACS Catal. 12, 14243–14263 (2022).
Judge, A. et al. Network of epistatic interactions in an enzyme active site revealed by large-scale deep mutational scanning. Proc. Natl Acad. Sci. USA 121, e2313513121 (2024).
Gelman, S., Fahlberg, S. A., Heinzelman, P., Romero, P. A. & Gitter, A. Neural networks to learn protein sequence–function relationships from deep mutational scanning data. Proc. Natl Acad. Sci. USA 118, e2104878118 (2021).
Kim, H. Y. & Kim, D. Prediction of mutation effects using a deep temporal convolutional network. Bioinformatics 36, 2047–2052 (2020).
Shanehsazzadeh, A., Belanger, D. & Dohan, D. Is transfer learning necessary for protein landscape prediction? In Proc. Machine Learning for Structural Biology Workshop in the Thirty-Fourth Annual Conference on Neural Information Processing Systems (NeurIPS) https://www.mlsb.io/papers/MLSB2020_Is_Transfer_Learning_Necessary.pdf (2020).
Yang, K. K., Lu, A. X. & Fusi, N. Convolutions are competitive with transformers for protein sequence pretraining. Cell System. 15, 286–294.e2 (2024).
Luo, Y. et al. ECNet is an evolutionary context-integrated deep learning framework for protein engineering. Nat. Commun. 12, 5743 (2021).
Hsu, C., Nisonoff, H., Fannjiang, C. & Listgarten, J. Learning protein fitness models from evolutionary and assay-labeled data. Nat. Biotechnol. 40, 1114–1122 (2022).
Sim, N.-L. et al. Sift web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 40, W452–W457 (2012).
Hopf, T. A. et al. Mutation effects predicted from sequence co-variation. Nat. Biotechnol. 35, 128–135 (2017).
Frazer, J. et al. Disease variant prediction with deep generative models of evolutionary data. Nature 599, 91–95 (2021).
Devlin, J., Chang, M.-W., Lee, K. & Toutanova, K. BERT: pre-training of deep bidirectional transformers for language understanding. In Proc. 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies 4171–4186 (Association for Computational Linguistics, 2019).
Floridi, L. & Chiriatti, M. GPT-3: its nature, scope, limits, and consequences. Minds Mach. 30, 681–694 (2020).
Rives, A. et al. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. Proc. Natl Acad. Sci. USA 118, e2016239118 (2021).
Hie, B., Zhong, E. D., Berger, B. & Bryson, B. Learning the language of viral evolution and escape. Science 371, 284–288 (2021).
Lin, Z. et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 379, 1123–1130 (2023).
Meier, J. et al. Language models enable zero-shot prediction of the effects of mutations on protein function. Adv. Neural Inf. Process. Syst. 34, 29287–29303 (2021).
Suzek, B. E. et al. Uniref clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 31, 926–932 (2015).
He, L. et al. Pre-training co-evolutionary protein representation via a pairwise masked language model. Preprint at https://arxiv.org/abs/2110.15527 (2021).
Dallago, C. et al. FLIP: benchmark tasks in fitness landscape inference for proteins. Preprint at bioRxiv https://doi.org/10.1101/2021.11.09.467890 (2021).
Hie, B. L. et al. Efficient evolution of human antibodies from general protein language models. Nat. Biotechnol. 41, 275–283 (2023).
Lu, H. et al. Machine learning-aided engineering of hydrolases for pet depolymerization. Nature 604, 662–667 (2022).
Hughes, D. & Andersson, D. I. Evolutionary trajectories to antibiotic resistance. Annu. Rev. Microbiol. 71, 579–596 (2017).
Wang, X., Zhang, H. & Chen, X. Drug resistance and combating drug resistance in cancer. Cancer Drug Resist. 2, 141 (2019).
Notin, P., Weitzman, R., Marks, D. & Gal, Y. ProteinNPT: improving protein property prediction and design with non-parametric transformers. Adv. Neural Inf. Process. Syst. 36, 33529–33563 (2023).
Zhao, J., Zhang, C. & Luo, Y. Contrastive fitness learning: reprogramming protein language models for low-n learning of protein fitness landscape. In Proc. 32nd International Conference on Pattern Recognition 470–474 (Springer, 2024).
Bryant, D. H. et al. Deep diversification of an aav capsid protein by machine learning. Nat. Biotechnol. 39, 691–696 (2021).
Jacquier, H. et al. Capturing the mutational landscape of the beta-lactamase TEM-1. Proc. Natl Acad. Sci. USA 110, 13067–13072 (2013).
Sarkisyan, K. S. et al. Local fitness landscape of the green fluorescent protein. Nature 533, 397–401 (2016).
Seuma, M., Faure, A. J., Badia, M., Lehner, B. & Bolognesi, B. The genetic landscape for amyloid beta fibril nucleation accurately discriminates familial Alzheimer’s disease mutations. eLife 10, e63364 (2021).
Faure, A. J. et al. Mapping the energetic and allosteric landscapes of protein binding domains. Nature 604, 175–183 (2022).
Araya, C. L. et al. A fundamental protein property, thermodynamic stability, revealed solely from large-scale measurements of protein function. Proc. Natl Acad. Sci. USA 109, 16858–16863 (2012).
Melamed, D., Young, D. L., Gamble, C. E., Miller, C. R. & Fields, S. Deep mutational scanning of an RRM domain of the Saccharomyces cerevisiae poly(A)-binding protein. RNA 19, 1537–1551 (2013).
Olson, C. A., Wu, N. C. & Sun, R. A comprehensive biophysical description of pairwise epistasis throughout an entire protein domain. Curr. Biol. 24, 2643–2651 (2014).
Starr, T. N. & Thornton, J. W. Epistasis in protein evolution. Protein Sci. 25, 1204–1218 (2016).
Sailer, Z. R. & Harms, M. J. High-order epistasis shapes evolutionary trajectories. PLoS Comput. Biol. 13, e1005541 (2017).
Naas, T. et al. Beta-lactamase database (BLDB)–structure and function. J. Enzyme Inhib. Med. Chem. 32, 917–919 (2017).
Stiffler, M. A., Hekstra, D. R. & Ranganathan, R. Evolvability as a function of purifying selection in TEM-1 β-lactamase. Cell 160, 882–892 (2015).
Weinreich, D. M., Delaney, N. F., DePristo, M. A. & Hartl, D. L. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312, 111–114 (2006).
Sinai, S. et al. AdaLead: a simple and robust adaptive greedy search algorithm for sequence design. Preprint at https://arxiv.org/abs/2010.02141 (2020).
Barrera, L. A. et al. Survey of variation in human transcription factors reveals prevalent DNA binding changes. Science 351, 1450–1454 (2016).
Chaudhury, S., Lyskov, S. & Gray, J. J. PyRosetta: a script-based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics 26, 689–691 (2010).
Ogden, P. J., Kelsic, E. D., Sinai, S. & Church, G. M. Comprehensive AAV capsid fitness landscape reveals a viral gene and enables machine-guided design. Science 366, 1139–1143 (2019).
Lorenz, R. et al. ViennaRNA package 2.0. Algorithms Mol. Biol. 6, 26 (2011).
Angermueller, C. et al. Model-based reinforcement learning for biological sequence design. In Proc. International Conference on Learning Representations https://openreview.net/forum?id=HklxbgBKvr (ICLR, 2020).
Brookes, D., Park, H. & Listgarten, J. Conditioning by adaptive sampling for robust design. In Proc. 36th International Conference on Machine Learning 773–782 (PMLR, 2019).
Hansen, N. The CMA evolution strategy: a tutorial. Preprint at https://arxiv.org/abs/1604.00772 (2016).
Kirjner, A. et al. Improving protein optimization with smoothed fitness landscapes. In Proc. International Conference on Learning Representations https://openreview.net/forum?id=rxlF2Zv8x0 (ICLR, 2024).
Wang, Y. et al. Self-play reinforcement learning guides protein engineering. Nat. Mach. Intell. 5, 845–860 (2023).
Wittmann, B. J., Yue, Y. & Arnold, F. H. Informed training set design enables efficient machine learning-assisted directed protein evolution. Cell Syst. 12, 1026–1045 (2021).
Qiu, Y., Hu, J. & Wei, G.-W. Cluster learning-assisted directed evolution. Nat. Comput. Sci. 1, 809–818 (2021).
De Visser, J. A. G. & Krug, J. Empirical fitness landscapes and the predictability of evolution. Nat. Rev. Genet. 15, 480–490 (2014).
Tokuriki, N. & Tawfik, D. S. Stability effects of mutations and protein evolvability. Curr. Opin. Struct. Biol. 19, 596–604 (2009).
Lenski, R. E., Barrick, J. E. & Ofria, C. Balancing robustness and evolvability. PLoS Biol. 4, e428 (2006).
Buel, G. R. & Walters, K. J. Can AlphaFold2 predict the impact of missense mutations on structure? Nat. Struct. Mol. Biol. 29, 1–2 (2022).
Wu, L. et al. SPRoBERTa: protein embedding learning with local fragment modeling. Brief. Bioinform. 23, bbac365 (2022).
Vaswani, A. et al. Attention is all you need. In Adv. Neural Inf. Process. Syst. 30, 5998–6008 (2017).
Hie, B., Zhong, E., Bryson, B. & Berger, B. Learning mutational semantics. Adv. Neural Inf. Process. Syst. 33, 9109–9121 (2020).
Bileschi, M. L. et al. Using deep learning to annotate the protein universe. Nat. Biotechnol. 40, 932–937 (2022).
Kulmanov, M. & Hoehndorf, R. DeepGOPlus: improved protein function prediction from sequence. Bioinformatics 36, 422–429 (2020).
Elnaggar, A. et al. ProtTrans: towards cracking the language of life’s code through self-supervised learning. IEEE Trans. Pattern Anal. Mach. Intell. 44, 7112–7127 (2021).
He, L., Deng, P. & Liu, G. μFormer encoder. figshare https://doi.org/10.6084/m9.figshare.26892355 (2024).
Schulman, J., Wolski, F., Dhariwal, P., Radford, A. & Klimov, O. Proximal policy optimization algorithms. Preprint at https://arxiv.org/abs/1707.06347 (2017).
OpenAI. Introducing ChatGPT; https://openai.com/blog/chatgpt
Ouyang, L. et al. Training language models to follow instructions with human feedback. In 36th Conference on Neural Information Processing Systems (NeurIPS 2022) https://proceedings.neurips.cc/paper_files/paper/2022/file/b1efde53be364a73914f58805a001731-Paper-Conference.pdf (2022).
Firnberg, E., Labonte, J. W., Gray, J. J. & Ostermeier, M. A comprehensive, high-resolution map of a gene’s fitness landscape. Mol. Biol. Evol. 31, 1581–1592 (2014).
Gonzalez, C. E. & Ostermeier, M. Pervasive pairwise intragenic epistasis among sequential mutations in tem-1 β-lactamase. J. Mol. Biol. 431, 1981–1992 (2019).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
He, L., Deng, P. & Liu, G. microsoft/Mu-Protein: MuProtein 0.1.1. Zenodo https://doi.org/10.5281/zenodo.15836168 (2025).
Acknowledgements
We gratefully acknowledge M. Ostermeier for generously providing detailed information on the plasmids pSkunk3-TEM-1 and pTS42, and for sharing the pTS42 plasmid itself. We also extend our thanks to T. Peng for developing the demonstration webpage and J. Bai for creating the graphic illustrations (the copyright for which is held by Microsoft as the work was completed during her employment). Last, we appreciate B. Kruft and U. Munir for their invaluable support in program management and coordination.
Author information
Authors and Affiliations
Contributions
Conceptualization: L.H., H.S. and P.D. Methodology and modelling: L.H., H.S., G.L., Z.Z., Y.J., F.J. and L.W. Data curation: L.H., H.S. and P.D. Result interpretation: P.D., H.L., L.H. and C.C. Writing—original draft: L.H., P.D., H.S. and G.L. Writing—review: H.L., T.Q. and C.C. Supervision: T.-Y.L. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Machine Intelligence thanks Yuchi Qiu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Ablation and analysis on μFormer components.
a) Ablation study to evaluate the importance of each component in μFormer. The change in performance after removing various components from the model relative to a full model is shown. Negative numbers (blue) indicate a loss of performance and positive numbers (red) indicate an improvement in performance. The last row displays the average performance change over 9 proteins. The plus/minus signs at the bottom indicate the presence/removal of the corresponding component. b) Spearman ρ statistics on 3 FLIP GB1 datasets of μFormer, ECNet, and their variants. ECNet w/ μFormer encoder replaces the language model in ECNet with μFormer’s language model. μFormer-S (Methods) is a variation with a model size similar to ECNet. 1-vs-rest: a train-test split where single-point mutants are used for training, and multi-point mutants are reserved for testing. 2-vs-rest: a train-test split where single- and double-point mutants are used for training, and all higher-order mutants are reserved for testing. 3-vs-rest: a train-test split where single-, double-, and triple-point mutants are used for training, and all higher-order mutants are reserved for testing. See Supplementary Notes for details.
Extended Data Fig. 2 Analysis of μProtein.
a) Performance of μFormer and Ridge on GB1 double mutants with varying training data size. Here, μFormer is a μFormer variation with a smaller supervised scorer module size (μFormer-SS). Training data ratio indicates the number of residues used for training versus the total number of amino acids in GB1. The training data size equals 209, 418, 627, 836, and 1045 for 20%, 40%, 60%, 80%, and 100%, respectively. All scores were evaluated on GB1 saturated double mutants (n=535,917). Center: mean. Error bands: standard deviation. Five experiments are performed for each setting with random selection on training data. b) Illustration of test data split, using a protein of 10 residues and the 40% setting as an example. 2/2 unseen: neither of the mutated residues in double mutants are seen by the model. 1/2 unseen: one and only one of the mutated residues in double mutants are seen by the model. c) Performance of μFormer and Ridge on different splits of GB1 double mutants. Training data split criteria are the same as in a). Center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range; points, outliers. Five experiments are performed for each setting with random selection on training data.
Supplementary information
Supplementary Information
Supplementary Notes 1–3, Figs. 1–11 and Tables 1–3.
Supplementary Data 1
Source data for Supplementary Figs. 1, 3, 5, 8, 9 and 11.
Supplementary Table 4
Source data for valid experiment results.
Source data
Source Data Figs. 2–6 and Extended Data Figs. 1 and 2
Source data for Figs. 2–6 and Extended Data Figs. 1 and 2.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sun, H., He, L., Deng, P. et al. Accelerating protein engineering with fitness landscape modelling and reinforcement learning. Nat Mach Intell 7, 1446–1460 (2025). https://doi.org/10.1038/s42256-025-01103-w
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s42256-025-01103-w
This article is cited by
-
Protein foundation models: a comprehensive survey
Science China Life Sciences (2026)
-
An integrated framework to accelerate protein design through mutagenesis
Nature Machine Intelligence (2025)