Abstract
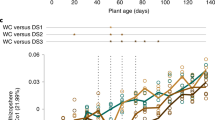
Drought stress impacts plant–microbe interactions, reshaping microbial community composition and biogeochemical cycling, thereby reducing crop productivity and threatening food security. However, the specific microbial responses and roles of plant-derived metabolites remain underexplored. Here we reveal that drought stress shifts the composition of wheat-associated microbiota across the phyllosphere, rhizosphere and root endosphere by favouring Actinobacteria and Ascomycota while depleting Proteobacteria and Basidiomycota. Targeted single-cell sorting and sequencing identified 21 active drought-tolerant bacteria (DTB) enriched in genes related to plant fitness and nutrient cycling. These DTB showed significant positive correlations with drought-enriched plant phytochemicals such as jasmonic acid and pipecolic acid. Moreover, the inoculation of synthetic community including four identified drought-tolerant taxa significantly stimulates the wheat growth under drought stress. A global exploration confirmed the widespread distribution of DTB, underscoring their promising potential to enhance crop resilience. This study provides new insights into drought-induced microbiome shifts and highlights microbial candidates for improving crop resilience in a changing climate.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All raw sequences were deposited in the National Center for Biotechnology Information Sequence Read Archive under accession number PRJNA1155890. All relevant metabolomic data, Raman data and R scripts, including those for microbial co-occurrence network construction and DTB phenotypic-level visualization, are available via GitHub at https://github.com/Lab-qlchen/Global-exploration-of-drought-tolerant-bacteria-in-wheat-rhizosphere. Source data are provided with this paper.
Change history
30 October 2025
A Correction to this paper has been published: https://doi.org/10.1038/s43016-025-01264-2
References
Dietz, K. J., Zörb, C. & Geilfus, C. M. Drought and crop yield. Plant Biol. 23, 881–893 (2021).
Vadez, V. et al. Crop traits and production under drought. Nat. Rev. Earth Environ. 5, 211–225 (2024).
Liu, H., Li, J. & Singh, B. K. Harnessing co-evolutionary interactions between plants and streptomyces to combat drought stress. Nat. Plants 10, 1159–1171 (2024).
Trivedi, P., Leach, J. E., Tringe, S. G., Sa, T. & Singh, B. K. Plant-microbiome interactions: from community assembly to plant health. Nat. Rev. Microbiol. 18, 607–621 (2020).
Liu, H., Brettell, L. E., Qiu, Z. & Singh, B. K. Microbiome-mediated stress resistance in plants. Trends Plant Sci. 25, 733–743 (2020).
Lebeis, S. L. et al. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349, 860–864 (2015).
Xu, L. et al. Drought delays development of the sorghum root microbiome and enriches for monoderm bacteria. Proc. Natl Acad. Sci. USA 115, E4284–E4293 (2018).
Veach, A. M. et al. Plant hosts modify belowground microbial community response to extreme drought. mSystems 5, e00092–20 (2020).
Duan, P. et al. Application of coronarin enhances maize drought tolerance by affecting interactions between rhizosphere fungal community and metabolites. Comput. Struct. Biotechnol. J. 21, 5273–5284 (2023).
Glick, B. R. Plant growth-promoting bacteria: mechanisms and applications. Scientifica 2012, 963401 (2012).
Barazani, O. & Friedman, J. Is IAA the major root growth factor secreted from plant-growth-mediating bacteria? J. Chem. Ecol. 25, 2397–2406 (1999).
Bei, Q. et al. Extreme summers impact cropland and grassland soil microbiomes. ISME J. 17, 1589–1600 (2023).
Lobell, D. B., Schlenker, W. & Costa-Roberts, J. Climate trends and global crop production since 1980. Science 333, 616–620 (2011).
Kahiluoto, H. et al. Decline in climate resilience of European wheat. Proc. Natl Acad. Sci. USA 116, 123–128 (2019).
van Dijk, A. I. J. M. et al. The millennium drought in southeast Australia (2001–2009): natural and human causes and implications for water resources, ecosystems, economy, and society. Water Resour. Res. 49, 1040–1057 (2013).
Li, H. Z. et al. Active antibiotic resistome in soils unraveled by single-cell isotope probing and targeted metagenomics. Proc. Natl Acad. Sci. USA 119, e2201473119 (2022).
No, J. H. et al. Raman-deuterium isotope probing and metagenomics reveal the drought tolerance of the soil microbiome and its promotion of plant growth. mSystems 7, e01249–01221 (2022).
Li, H. Z. et al. Single-cell exploration of active phosphate-solubilizing bacteria across diverse soil matrices for sustainable phosphorus management. Nat. Food. 5, 673–683 (2024).
Li, M., Xu, J., Romero-Gonzalez, M., Banwart, S. A. & Huang, W. E. Single cell raman spectroscopy for cell sorting and imaging. Curr. Opin. Biotechnol. 23, 56–63 (2012).
Yin, J. et al. Future socio-ecosystem productivity threatened by compound drought–heatwave events. Nat. Sustain. 6, 259–272 (2023).
Anjum, S. A. et al. Morphological, physiological and biochemical responses of plants to drought stress. Afr. J. Agric. Res. 6, 2026–2032 (2011).
Ahmad, Z. et al. Physiological responses of wheat to drought stress and its mitigation approaches. Acta Physiol. Plant. 40, 80 (2018).
Ruehr, N. K. et al. Drought effects on allocation of recent carbon: from beech leaves to soil CO2 efflux. New Phytol. 184, 950–961 (2009).
Yadav, B., Jogawat, A., Rahman, M. S. & Narayan, O. P. Secondary metabolites in the drought stress tolerance of crop plants: a review. Gene Rep. 23, 101040 (2021).
Vergara-Diaz, O. et al. Metabolome profiling supports the key role of the spike in wheat yield performance. Cells 9, 1025 (2020).
Wang, P. et al. A novel role of pipecolic acid biosynthetic pathway in drought tolerance through the antioxidant system in tomato. Antioxidants 10, 1923 (2021).
Hu, L. et al. Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 9, 2738 (2018).
Yu, P. et al. Plant flavones enrich rhizosphere oxalobacteraceae to improve maize performance under nitrogen deprivation. Nat. Plants 7, 481–499 (2021).
Cushnie, T. P. T., Cushnie, B. & Lamb, A. J. Alkaloids: an overview of their antibacterial, antibiotic-enhancing and antivirulence activities. Int. J. Antimicrob. Agents 44, 377–386 (2014).
Carvalhais, L. C. et al. Linking jasmonic acid signaling, root exudates, and rhizosphere microbiomes. Mol. Plant-Microbe Interact. 28, 1049–1058 (2015).
Wang, Y., Mostafa, S., Zeng, W. & Jin, B. Function and mechanism of jasmonic acid in plant responses to abiotic and biotic stresses. Int. J. Mol. Sci. 22, 8568 (2021).
Arellano-Wattenbarger, G. L., Montiel, S., Aguirre-Von-Wobeser, E., de la Torre, M. & Rocha, J. Contribution of seed-endophytic bacteria to drought tolerance in early developmental stages of native maize landraces from arid milpas. Plant Soil 500, 213–232 (2023).
Wink, M. in Modern Alkaloids: Structure, Isolation, Synthesis and Biology (eds Fattorusso, E. & Taglialatela-Scafati, O.) 1–24 (Wiley-VCH, 2008).
de Vries, F. T., Griffiths, R. I., Knight, C. G., Nicolitch, O. & Williams, A. Harnessing rhizosphere microbiomes for drought-resilient crop production. Science 368, 270–274 (2020).
Haskett, T. L., Tkacz, A. & Poole, P. S. Engineering rhizobacteria for sustainable agriculture. ISME J. 15, 949–964 (2021).
Fan, K. et al. Biodiversity of key-stone phylotypes determines crop production in a 4-decade fertilization experiment. ISME J. 15, 550–561 (2021).
Jiao, S., Lu, Y. & Wei, G. Soil multitrophic network complexity enhances the link between biodiversity and multifunctionality in agricultural systems. Glob. Change Biol. 28, 140–153 (2021).
Vilchez, J. I., Garcia-Fontana, C., Roman-Naranjo, D., Gonzalez-Lopez, J. & Manzanera, M. Plant drought tolerance enhancement by trehalose production of desiccation-tolerant microorganisms. Front. Microbiol. 7, 1577 (2016).
Sunita, K., Mishra, I., Mishra, J., Prakash, J. & Arora, N. K. Secondary metabolites from halotolerant plant growth promoting rhizobacteria for ameliorating salinity stress in plants. Front. Microbiol. 11, 567768 (2020).
Moreno, J. C. & Al-Babili, S. Are carotenoids the true colors of crop improvement? New Phytol. 237, 1946–1950 (2023).
Xu, L. et al. Genome-resolved metagenomics reveals role of iron metabolism in drought-induced rhizosphere microbiome dynamics. Nat. Commun. 12, 3209 (2021).
Wolińska, A. & Stępniewska, Z. Dehydrogenase activity in the soil environment. Dehydrogenases 10, 183–210 (2012).
Chen, C. et al. Pantoea alhagi, a novel endophytic bacterium with ability to improve growth and drought tolerance in wheat. Sci. Rep. 7, 41564 (2017).
Kour D. et al. in Plant Growth Promoting Rhizobacteria for Sustainable Stress Management: Vol. 1: Rhizobacteria in Abiotic Stress Management (eds Sayyed R. Z. et al.) 255–308 (Springer, 2019).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. Fastp: an ultra-fast all-in-one fastq preprocessor. Bioinformatics 34, i884–i890 (2018).
Li, D., Liu, C. M., Luo, R., Sadakane, K. & Lam, T. W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de bruijn graph. Bioinformatics 31, 1674–1676 (2015).
Noguchi, H., Park, J. & Takagi, T. MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 34, 5623–5630 (2006).
Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012).
Li, R., Li, Y., Kristiansen, K. & Wang, J. SOAP: short oligonucleotide alignment program. Bioinformatics 24, 713–714 (2008).
Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015).
Ma, B. et al. A genomic catalogue of soil microbiomes boosts mining of biodiversity and genetic resources. Nat. Commun. 14, 7318 (2023).
Blin, K. et al. antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 49, W29–W35 (2021).
Wang, Z. et al. Root hair developmental regulators orchestrate drought triggered microbiome changes and the interaction with beneficial rhizobiaceae. Nat. Commun. 15, 10068 (2024).
Rohart, F., Gautier, B., Singh, A. & Lê Cao, K.-A. Mixomics: an R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 13, e1005752 (2017).
Wickham, H., Chang, W. & Wickham, M. H. ggplot2: Create elegant data visualisations using the grammar of graphics. R package version 2 (2016); https://CRAN.R-project.org/package=ggplot2
Oksanen J. et al. vegan: Community ecology package. R package version 2 (2018); https://CRAN.R-project.org/package=vegan
Roberts, D. W. & Roberts, M. D. W. labdsv: Ordination and multivariate analysis for ecology. R package version 1.8‑0 (2016); https://CRAN.R-project.org/package=labdsv
Acknowledgements
We acknowledge the funds of the Natural Science Foundation of Fujian Province (grant number 2023J02029 to Q.-L.C), National Natural Science Foundation of China (grant number 42207143 to Q.X.) and the fellowship of China Postdoctoral Science Foundation (grant number BX2021293 to Q.X.).
Author information
Authors and Affiliations
Contributions
Q.X., Q.-L.C. and Y.-G.Z. conceived and designed the research. Q.X. and K.Y. performed the experiments and analysed the data with the help of B.M. and C.-Y.L. Q.X. and K.Y. wrote the paper with the assistance of all the co-authors. All authors read and approved the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Food thanks Josep Peñuelas and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary results (Texts 1–3), methods (Texts 4–6) and Figs. 1–20.
Source data
Source Data Fig. 1
Wheat metabolome data and the statistical source data for Fig. 1e,f.
Source Data Fig. 3
Statistical source data for Fig. 3a (normalized C–D ratio, DTB percentage and drought-tolerant level) and Fig. 3b (active DTB with normalized C–D ratios > 1.6).
Source Data Fig. 4
Statistical source data for Fig. 4c, showing the correlations between DTB and upregulated phytochemicals in leaf and root tissues.
Source Data Fig. 5
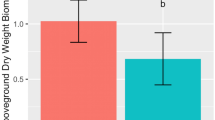
Statistical source data for Fig. 5a,c, showing the effect of DTB on wheat growth parameters under drought.
Source Data Fig. 6
Statistical source data for Fig. 6a,b, including the data of DTB abundance, Shannon index of DT, standardized precipitation evapotranspiration index, and the latitude and longitude of samples.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xiang, Q., Yang, K., Cui, L. et al. Global exploration of drought-tolerant bacteria in the wheat rhizosphere reveals microbiota shifts and functional taxa enhancing plant resilience. Nat Food 6, 1054–1067 (2025). https://doi.org/10.1038/s43016-025-01248-2
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s43016-025-01248-2
This article is cited by
-
Drought-tolerant rhizobacteria boost crop resilience
Nature Food (2025)