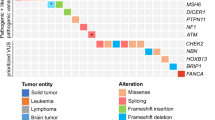
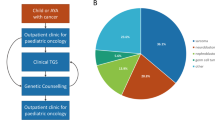
Abstract
The spectrum of germline predisposition in pediatric cancer continues to be realized. Here we report 751 patients with solid tumors who underwent prospective matched tumor–normal DNA sequencing with downstream clinical use (ClinicalTrials.gov NCT01775072). Germline pathogenic and likely pathogenic (P/LP) variants were reported. One or more P/LP variants were found in 18% (138/751) of individuals when including variants in low-, moderate- and high-penetrance dominant or recessive genes or in 13% (99/751) of individuals in moderate- and high-penetrance dominant genes. Thirty-four percent of high- or moderate-penetrance variants were unexpected based on the patient’s diagnosis and previous history. Seventy-six percent of patients with positive results completed a clinical genetics visit, and 21% had at least one relative undergo cascade testing as a result of this testing. Clinical actionability additionally included screening, risk reduction in relatives, reproductive use and targeted therapies. Germline testing should be considered for all children with cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
Deidentified clinical and molecular data for all patients reported in this study are available in Supplementary Table 1. P/LP variants reported here have been submitted to ClinVar with submission number SUB8689141.
References
Howlader, N. et al. (eds.) SEER Cancer Statistics Review, 1975–2014 (National Cancer Institute, 2017).
Zhang, J. et al. Germline mutations in predisposition genes in pediatric cancer. N. Engl. J. Med. 373, 2336–2346 (2015).
Mody, R. J. et al. Integrative clinical sequencing in the management of refractory or relapsed cancer in youth. JAMA 314, 913–925 (2015).
Harris, M. H. et al. Multicenter feasibility study of tumor molecular profiling to inform therapeutic decisions in advanced pediatric solid tumors: the individualized cancer therapy (iCat) study. JAMA Oncol. 2, 608–615 (2016).
Oberg, J. A. et al. Implementation of next generation sequencing into pediatric hematology–oncology practice: moving beyond actionable alterations. Genome Med. 8, 133 (2016).
Parsons, D. W. et al. Diagnostic yield of clinical tumor and germline whole-exome sequencing for children with solid tumors. JAMA Oncol. 2, 616–624 (2016).
Wong, M. et al. Whole genome, transcriptome and methylome profiling enhances actionable target discovery in high-risk pediatric cancer. Nat. Med. 26, 1742–1753 (2020).
Walsh, M. et al. in Abeloff’s Clinical Oncology 6th edn (eds Niederhuber, J. E. et al.) Ch. 13 (Elsevier, 2020).
Holmfeldt, L. et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat. Genet. 45, 242–252 (2013).
Scollon, S. et al. A comprehensive review of pediatric tumors and associated cancer predisposition syndromes. J. Genet. Couns. 26, 387–434 (2017).
Suerink, M. et al. Constitutional mismatch repair deficiency as a differential diagnosis of neurofibromatosis type 1: consensus guidelines for testing a child without malignancy. J. Med. Genet. 56, 53–62 (2019).
Gault, M. D. et al. Germline SDHA mutations in children and adults with cancer. Cold Spring Harb. Mol. Case Stud. 4, a002584 (2018).
Oak, N. et al. Ancestry-specific predisposing germline variants in cancer. Genome Med. 12, 51 (2020).
Grzymski, J. J. et al. Population genetic screening efficiently identifies carriers of autosomal dominant diseases. Nat. Med. 26, 1235–1239 (2020).
Walsh, M.F. et al. Germline BRCA2 mutations detected in pediatric sequencing studies impact parents’ evaluation and care. Cold Spring Harb. Mol. Case Stud. 3, a001925 (2017).
Offit, K. et al. Cascading after peridiagnostic cancer genetic testing: an alternative to population-based screening. J. Clin. Oncol. 38, 1398–1408 (2020).
Le, D. T. et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 372, 2509–2520 (2015).
Moore, K. et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N. Engl. J. Med. 379, 2495–2505 (2018).
Bouffet, E. et al. Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J. Clin. Oncol. 34, 2206–2211 (2016).
Gross, A. M. et al. Selumetinib in children with inoperable plexiform neurofibromas. N. Engl. J. Med. 382, 1430–1442 (2020).
Fangusaro, J. et al. Selumetinib in paediatric patients with BRAF-aberrant or neurofibromatosis type 1-associated recurrent, refractory, or progressive low-grade glioma: a multicentre, phase 2 trial. Lancet Oncol. 20, 1011–1022 (2019).
van Vuurden, D. G. et al. PARP inhibition sensitizes childhood high grade glioma, medulloblastoma and ependymoma to radiation. Oncotarget 2, 984–996 (2011).
Mandelker, D. et al. Mutation detection in patients with advanced cancer by universal sequencing of cancer-related genes in tumor and normal DNA vs guideline-based germline testing. JAMA 318, 825–835 (2017).
Walsh, M. F. et al. Integrating somatic variant data and biomarkers for germline variant classification in cancer predisposition genes. Hum. Mutat. 39, 1542–1552 (2018).
Kratz, C. P. et al. Cancer screening recommendations for individuals with Li–Fraumeni syndrome. Clin. Cancer Res. 23, e38–e45 (2017).
Rednam, S. P. et al. Von Hippel–Lindau and hereditary pheochromocytoma/paraganglioma syndromes: clinical features, genetics, and surveillance recommendations in childhood. Clin. Cancer Res. 23, e68–e75 (2017).
Fiala, E. M. et al. 11p15.5 epimutations in children with Wilms tumor and hepatoblastoma detected in peripheral blood. Cancer 126, 3114–3121 (2020).
Benayed, R. et al. High yield of RNA sequencing for targetable kinase fusions in lung adenocarcinomas with no mitogenic driver alteration detected by DNA sequencing and low tumor mutation burden. Clin. Cancer Res. 25, 4712–4722 (2019).
Karam, R. et al. Assessment of diagnostic outcomes of RNA genetic testing for hereditary cancer. JAMA Netw. Open 2, e1913900 (2019).
Knapke, S. et al. Hereditary cancer risk assessment in a pediatric oncology follow-up clinic. Pediatr. Blood Cancer 58, 85–89 (2012).
Mirabello, L. et al. Frequency of pathogenic germline variants in cancer-susceptibility genes in patients with osteosarcoma. JAMA Oncol. 6, 724–734 (2020).
Qin, N. et al. Pathogenic germline mutations in DNA repair genes in combination with cancer treatment exposures and risk of subsequent neoplasms among long-term survivors of childhood cancer. J. Clin. Oncol. 38, 2728–2740 (2020).
Sharaf, R. N. et al. Uptake of genetic testing by relatives of Lynch syndrome probands: a systematic review. Clin. Gastroenterol. Hepatol. 11, 1093–1100 (2013).
Courtney, E. et al. Impact of free cancer predisposition cascade genetic testing on uptake in Singapore. NPJ Genom. Med. 4, 22 (2019).
Frey, M. K. et al. Prospective feasibility trial of a novel strategy of facilitated cascade genetic testing using telephone counseling. J. Clin. Oncol. 38, 1389–1397 (2020).
Srinivasan, S. et al. Stakeholder perspectives on overcoming barriers to cascade testing in Lynch syndrome: a qualitative study. Cancer Prev. Res. 13, 1037–1046 (2020).
Tak, C. R. et al. Cost-effectiveness of early cancer surveillance for patients with Li–Fraumeni syndrome. Pediatr. Blood Cancer 66, e27629 (2019).
Cheng, D. T. et al. Memorial Sloan Kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J. Mol. Diagn. 17, 251–264 (2015).
Zehir, A. et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 23, 703–713 (2017).
Cheng, D. T. et al. Comprehensive detection of germline variants by MSK-IMPACT, a clinical diagnostic platform for solid tumor molecular oncology and concurrent cancer predisposition testing. BMC Med. Genomics 10, 33 (2017).
Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–424 (2015).
Tung, N. et al. Counselling framework for moderate-penetrance cancer-susceptibility mutations. Nat. Rev. Clin. Oncol. 13, 581–588 (2016).
Shen, R. & Seshan, V. E. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 44, e131 (2016).
Jonsson, P. et al. Tumour lineage shapes BRCA-mediated phenotypes. Nature 571, 576–579 (2019).
Niu, B. et al. MSIsensor: microsatellite instability detection using paired tumor–normal sequence data. Bioinformatics 30, 1015–1016 (2014).
Acknowledgements
This work was funded in part by the Robert and Kate Niehaus Center for Inherited Cancer Genomics, the Pediatric Translational Medicine Program, the Marie-Josée and Henry R. Kravis Center for Molecular Oncology and the National Cancer Institute Cancer Center Core grant number P30-CA008748.
Author information
Authors and Affiliations
Contributions
M.F.W., E.M.F. and A.Z. conceptualized and designed the study. E.M.F., G.J., A. Mauguen, J.A.K., N.B., Y.K., M.H.F., A. Maio, E.E.S.-M., M. Sheehan, A.G.A., A.L., M.I.C., K.C., S. Murkherjee, E.K.S., T.T., J.G.B., P.A.M., L.W., F.S.D.C., N.-K.C., E.B., A.K., M.O., J.H.F., I.J.D., Y.K., S.G., S.F.S., C.J.F., M. Sulis, M.K., S. Modak, T.E.H., S.R., C.Y., S.J., J.V., S.T., D.N.F., Z.K.S., M.R., M.F.B., N.S., M.L., R.J.O.R., D.H.A., O.C.-B., L.Z., D.M., N.N.S., A.L.K., K.O., A.Z. and M.F.W. acquired, analyzed and interpreted the data. M.F.W. and E.M.F. drafted the manuscript. G.J., A. Mauguen, J.A.K., N.B., Y.K., M.H.F., A. Maio, E.E.S.-M., M. Sheehan, A.G.A., A.L., M.I.C., K.C., S. Murkherjee, E.K.S., T.T., J.G.B., P.A.M., L.W., F.S.D.C., N.-K.C., E.B., A.K., M.O., J.H.F., I.J.D., Y.K., S.G., S.F.S., C.J.F., M. Sulis, M.K., S. Modak, J.T.G., T.E.H., S.R., C.Y., S.J., J.V., S.T., D.N.F., Z.K.S., M.R., M.F.B., N.S., M.L., R.J.O.R., D.H.A., O.C.-B., L.Z., D.M., N.N.S., A.L.K., K.O. and A.Z. critically revised the manuscript for important intellectual content. A. Mauguen performed the statistical analysis. M.F.W., A.L.K., K.O. and M.L. obtained funding. E.M.F., J.A.K., N.B., Y.K., M.H.F., A. Maio, E.E.S.-M., M. Sheehan and A.G.A. performed administrative, technical or material support. M.F.W., A.Z., A.L.K. and K.O. supervised the study.
Corresponding authors
Ethics declarations
Competing interests
P.A.M.: Takeda, uncompensated speaking; Amgen, stock ownership; Ipsen, mock ODAC panel; Salarius, mock ODAC panel; P.A.M.’s spouse: Boehringer Ingelheim, consulting; Genentech, speakers’ bureau; Eastern Pulmonary Society. N.K.C. holds ownership interest/equity in Y-mAbs Therapeutics, Inc., reports receiving commercial research grants from Y-mAbs Therapeutics, Inc., and Abpro Labs, Inc., holding ownership interest/equity in Abpro Labs and owning stock options in Eureka Therapeutics, Inc. N.K.C. is a scientific advisory board member of Abpro Labs and Eureka Therapeutics. N.K.C. is the inventor and owner of issued patents licensed by MSK to Y-mAbs Therapeutics, Biotec Pharmacon and Abpro Labs. I.J.D. has done consulting and advisory board activities with Apexigen, AstraZeneca, Bayer, Bristol-Myers Squibb/Celgene, Fennec and Roche. I.J.D. has served as an institutional PI for pharma-sponsored clinical trials at MSKCC from Bristol-Myers Squibb, Genentech and Novartis. M.K. has consultant agreements with AstraZeneca, Bayer, CereXis, QED Therapeutics and Recursion Pharma (personal fees received). S. Modak. is a consultant to Y-mAbs Therapeutics, Inc., Illumina, Inc., and Progenics Pharmaceuticals. Z.K.S.’s immediate family member serves as a consultant for Allergan, Adverum Biotechnologies, Alimera Sciences, Fortress Biotech, Genentech/Roche, Novartis, Optos, Regeneron, Regenxbio and Spark Therapeutics.
Additional information
Peer review information Nature Cancer thanks Todd Druley, Heather Hampel and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Fiala, E.M., Jayakumaran, G., Mauguen, A. et al. Prospective pan-cancer germline testing using MSK-IMPACT informs clinical translation in 751 patients with pediatric solid tumors. Nat Cancer 2, 357–365 (2021). https://doi.org/10.1038/s43018-021-00172-1
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s43018-021-00172-1
This article is cited by
-
Evaluating the potential and limitations of nanopore adaptive sampling for targeted transcriptome sequencing
Genome Biology (2025)
-
Extended spectrum of cancers in PTEN hamartoma tumor syndrome
npj Precision Oncology (2025)
-
A coordinated multidisciplinary model of care is needed for child and family centered care in pediatric genetic cancer risk services: a scoping review
Familial Cancer (2025)
-
Genetic landscape of cancer: mechanisms, key genes, and therapeutic implications
Clinical and Translational Oncology (2025)
-
Humangenetische Diagnostik bei onkologischen Erkrankungen
InFo Hämatologie + Onkologie (2025)