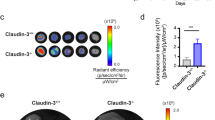
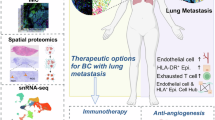
Abstract
Carcinomas are associated with metastasis to specific organs while sparing others. Breast cancer presents with lung metastasis but rarely kidney metastasis. Using this difference as an example, we queried the mechanism(s) behind the proclivity for organ-specific metastasis. We used spontaneous and implant models of metastatic mammary carcinoma coupled with inflammatory tissue fibrosis, single-cell sequencing analyses and functional studies to unravel the causal determinants of organ-specific metastasis. Here we show that lung metastasis is facilitated by angiopoietin 2 (Ang2)-mediated suppression of lung-specific endothelial tight junction protein Claudin 5, which is augmented by the inflammatory fibrotic microenvironment and prevented by anti-Ang2 blocking antibodies, while kidney metastasis is prevented by non-Ang2-responsive Claudins 2 and 10. Suppression of Claudins 2 and 10 was sufficient to induce the emergence of kidney metastasis. This study illustrates the influence of organ-specific vascular heterogeneity in determining organotropic metastasis, independent of cancer cell-intrinsic mechanisms.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Microarray and RNA-seq or scRNA-seq data that support the findings of this study were deposited to the GEO under accession code GSE199178. Previously published microarray data that were reanalyzed herein are available as follows: scRNA-seq of murine ECs (ArrayExpress: E-MTAB-8077)34, murine and human lung tissue (Synapse: syn21560406; https://www.synapse.org/#!Synapse:syn21560406)35, murine kidney tissue (GEO: GSE157079)36, human kidney tissue (Human Cell Atlas Data Portal; https://explore.data.humancellatlas.org/projects/abe1a013-af7a-45ed-8c26-f3793c24a1f4)37, murine brain tissue (ArrayExpress: E-MTAB-8077)34 and human brain tissue (GEO: GSE162631)61. All other data supporting the findings of this study are available from the corresponding author on reasonable request. No custom code was generated in the course of this study. Source data are provided with this paper.
References
Chaffer, C. L. & Weinberg, R. A. A perspective on cancer cell metastasis. Science 331, 1559–1564 (2011).
Kennecke, H. et al. Metastatic behavior of breast cancer subtypes. J. Clin. Oncol. 28, 3271–3277 (2010).
Paget, S. The distribution of secondary growths in cancer of the breast. Cancer Metastasis Rev. 8, 98–101 (1989).
Kaplan, R. N., Rafii, S. & Lyden, D. Preparing the ‘soil’: the premetastatic niche. Cancer Res. 66, 11089–11093 (2006).
Gao, Y. et al. Metastasis organotropism: redefining the congenial soil. Dev. Cell 49, 375–391 (2019).
Rezaie, J. et al. Tumor-derived extracellular vesicles: the metastatic organotropism drivers. Life Sci. 289, 120216 (2022).
Wortzel, I., Dror, S., Kenific, C. M. & Lyden, D. Exosome-mediated metastasis: communication from a distance. Dev. Cell 49, 347–360 (2019).
Peinado, H. et al. Pre-metastatic niches: organ-specific homes for metastases. Nat. Rev. Cancer 17, 302–317 (2017).
Reymond, N., d’Agua, B. B. & Ridley, A. J. Crossing the endothelial barrier during metastasis. Nat. Rev. Cancer 13, 858–870 (2013).
Augustin, H. G. & Koh, G. Y.Organotypic vasculature: from descriptive heterogeneity to functional pathophysiology. Science 357, eaal2379 (2017).
Ghajar, C. M. et al. The perivascular niche regulates breast tumour dormancy. Nat. Cell Biol. 15, 807–817 (2013).
Cooke, V. G. et al. Pericyte depletion results in hypoxia-associated epithelial-to-mesenchymal transition and metastasis mediated by Met signaling pathway. Cancer Cell 21, 66–81 (2012).
Kim, J. et al. Heterogeneous perivascular cell coverage affects breast cancer metastasis and response to chemotherapy. JCI Insight 1, e90733 (2016).
Rybinski, B., Franco-Barraza, J. & Cukierman, E. The wound healing, chronic fibrosis, and cancer progression triad. Physiol. Genomics 46, 223–244 (2014).
Chen, Y. et al. Dual reporter genetic mouse models of pancreatic cancer identify an epithelial-to-mesenchymal transition-independent metastasis program. EMBO Mol. Med. 10, e9085 (2018).
Becker, L. M. et al. Epigenetic reprogramming of cancer-associated fibroblasts deregulates glucose metabolism and facilitates progression of breast cancer. Cell Rep. 31, 107701 (2020).
Carstens, J. L. et al. Stabilized epithelial phenotype of cancer cells in primary tumors leads to increased colonization of liver metastasis in pancreatic cancer. Cell Rep. 35, 108990 (2021).
Piersma, B., Hayward, M. K. & Weaver, V. M. Fibrosis and cancer: a strained relationship. Biochim. Biophys. Acta Rev. Cancer 1873, 188356 (2020).
Chandler, C., Liu, T., Buckanovich, R. & Coffman, L. G. The double edge sword of fibrosis in cancer. Transl. Res. 209, 55–67 (2019).
Cox, T. R. et al. LOX-mediated collagen crosslinking is responsible for fibrosis-enhanced metastasis. Cancer Res. 73, 1721–1732 (2013).
Zhou, C. et al. Metastases to the kidney: a comprehensive analysis of 151 patients from a tertiary referral centre. BJU Int. 117, 775–782 (2016).
Cazacu, S. M. et al. Metastases to the kidney: a case report and review of the literature. Curr. Health Sci. J. 46, 80–89 (2020).
Benest, A. V. et al. Angiopoietin-2 is critical for cytokine-induced vascular leakage. PLoS ONE 8, e70459 (2013).
Holopainen, T. et al. Effects of angiopoietin-2-blocking antibody on endothelial cell–cell junctions and lung metastasis. J. Natl Cancer Inst. 104, 461–475 (2012).
Keskin, D. et al. Targeting vascular pericytes in hypoxic tumors increases lung metastasis via angiopoietin-2. Cell Rep. 10, 1066–1081 (2015).
Gengenbacher, N. et al. Timed Ang2-targeted therapy identifies the angiopoietin–Tie pathway as key regulator of fatal lymphogenous metastasis. Cancer Discov. 11, 424–445 (2021).
Park, J. S. et al. Normalization of tumor vessels by Tie2 activation and Ang2 inhibition enhances drug delivery and produces a favorable tumor microenvironment. Cancer Cell 31, 157–158 (2017).
Srivastava, K. et al. Postsurgical adjuvant tumor therapy by combining anti-angiopoietin-2 and metronomic chemotherapy limits metastatic growth. Cancer Cell 26, 880–895 (2014).
Rigamonti, N. & De Palma, M. A role for angiopoietin-2 in organ-specific metastasis. Cell Rep. 4, 621–623 (2013).
Li, P., He, Q., Luo, C. & Qian, L. Diagnostic and prognostic potential of serum angiopoietin-2 expression in human breast cancer. Int. J. Clin. Exp. Pathol. 8, 660–664 (2015).
Tiainen, L. et al. High baseline Tie1 level predicts poor survival in metastatic breast cancer. BMC Cancer 19, 732 (2019).
Avraham, H. K. et al. Angiopoietin-2 mediates blood–brain barrier impairment and colonization of triple-negative breast cancer cells in brain. J. Pathol. 232, 369–381 (2014).
Zihni, C., Mills, C., Matter, K. & Balda, M. S. Tight junctions: from simple barriers to multifunctional molecular gates. Nat. Rev. Mol. Cell Biol. 17, 564–580 (2016).
Kalucka, J. et al. Single-cell transcriptome atlas of murine endothelial cells. Cell 180, 764–779 (2020).
Travaglini, K. J. et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature 587, 619–625 (2020).
Miao, Z. et al. Single cell regulatory landscape of the mouse kidney highlights cellular differentiation programs and disease targets. Nat. Commun. 12, 2277 (2021).
Stewart, B. J. et al. Spatiotemporal immune zonation of the human kidney. Science 365, 1461–1466 (2019).
Van Itallie, C. M. et al. Two splice variants of claudin-10 in the kidney create paracellular pores with different ion selectivities. Am. J. Physiol. Renal Physiol. 291, F1288–F1299 (2006).
Taddei, A. et al. Endothelial adherens junctions control tight junctions by VE-cadherin-mediated upregulation of claudin-5. Nat. Cell Biol. 10, 923–934 (2008).
Kim, M. et al. Opposing actions of angiopoietin-2 on Tie2 signaling and FOXO1 activation. J. Clin. Invest. 126, 3511–3525 (2016).
Zhou, X. et al. Acute kidney injury instigates malignant renal cell carcinoma via CXCR2 in mice with inactivated Trp53 and Pten in proximal tubular kidney epithelial cells. Cancer Res. 81, 2690–2702 (2021).
Fidler, I. J. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat. Rev. Cancer 3, 453–458 (2003).
Hart, I. R. & Fidler, I. J. Role of organ selectivity in the determination of metastatic patterns of B-16 melanoma. Cancer Res. 40, 2281–2287 (1980).
Hongu, T. et al. Perivascular tenascin C triggers sequential activation of macrophages and endothelial cells to generate a pro-metastatic vascular niche in the lungs. Nat. Cancer 3, 486–504 (2022).
Singhal, M. et al. Temporal multi-omics identifies LRG1 as a vascular niche instructor of metastasis. Sci. Transl. Med. 13, eabe6805 (2021).
Chang, F. C. et al. Angiopoietin-2 inhibition attenuates kidney fibrosis by hindering chemokine C–C motif ligand 2 expression and apoptosis of endothelial cells. Kidney Int. 102, 780–797 (2022).
Parikh, S. M. et al. Excess circulating angiopoietin-2 may contribute to pulmonary vascular leak in sepsis in humans. PLoS Med. 3, e46 (2006).
Uehara, M. et al. Impact of angiopoietin-1 and -2 on clinical course of idiopathic pulmonary fibrosis. Respir. Med. 114, 18–26 (2016).
Gallagher, D. C. et al. Angiopoietin 2 is a potential mediator of high-dose interleukin 2-induced vascular leak. Clin. Cancer Res. 13, 2115–2120 (2007).
Hashimoto, T. & Pittet, J. F. Angiopoietin-2: modulator of vascular permeability in acute lung injury? PLoS Med. 3, e113 (2006).
Huang, H. H., Bhat, A., Woodnutt, G. & Lappe, R. Targeting the ANGPT–TIE2 pathway in malignancy. Nat. Rev. Cancer 10, 575–585 (2010).
Sfiligoi, C. et al. Angiopoietin-2 expression in breast cancer correlates with lymph node invasion and short survival. Int. J. Cancer 103, 466–474 (2003).
Pauta, M. et al. Overexpression of angiopoietin-2 in rats and patients with liver fibrosis. Therapeutic consequences of its inhibition. Liver Int. 35, 1383–1392 (2015).
Lovisa, S. et al. Epithelial-to-mesenchymal transition induces cell cycle arrest and parenchymal damage in renal fibrosis. Nat. Med. 21, 998 (2015).
McAndrews, K. M. et al. Dermal αSMA+ myofibroblasts orchestrate skin wound repair via beta1 integrin and independent of type I collagen production. EMBO J. 41, e109470 (2022).
Lin, E. Y. et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am. J. Pathol. 163, 2113–2126 (2003).
Kaner, R. J. et al. Lung overexpression of the vascular endothelial growth factor gene induces pulmonary edema. Am. J. Respir. Cell Mol. Biol. 22, 657–664 (2000).
Liao, Y., Wang, J., Jaehnig, E. J., Shi, Z. & Zhang, B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 47, W199–W205 (2019).
Benjamini, Y., Drai, D., Elmer, G., Kafkafi, N. & Golani, I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 125, 279–284 (2001).
McAndrews, K. M. et al. Heterogeneous antibodies against SARS-CoV-2 spike receptor binding domain and nucleocapsid with implications for COVID-19 immunity. JCI Insight 5, e142386 (2020).
Xie, Y. et al. Key molecular alterations in endothelial cells in human glioblastoma uncovered through single-cell RNA sequencing. JCI Insight 6, e150861 (2021).
Garcia-Recio, S. et al. Multiomics in primary and metastatic breast tumors from the AURORA US network finds microenvironment and epigenetic drivers of metastasis. Nat. Cancer 4, 128–147 (2023).
Acknowledgements
We thank I. J. Fidler for his guidance and discussion of our results. We thank M. Thomas, J. Muller and Y. Kienast (Roche, Germany) for providing the anti-Ang2 neutralizing antibody. We thank M. Kirtley, P. Phillips, K. Vadnagara, D. Lundy and L. Gibson for technical assistance. This work was primarily supported by National Cancer Instute (NCI) grant R01CA252729. R.K. was supported by the CPRIT RP200612 grant, the Metastasis Research Center at MDACC, research funding support from Champalimaud Foundation (Lisbon, Portugal), the Albert and Margaret Alkek Foundation and Distinguished University Chair supported by the Sid W. Richardson Foundation. V.S.L. was in part supported by the NIH NCI Cancer Center Support Grant New Faculty Award P30CA016672. STR validation was performed by the Cytogenetics and Cell Authentication Core at MDACC. The general operations of the Advanced Technology Genomics Core, Flow Cytometry and Cellular Imaging Core and Small Animal Imaging Facility at MDACC was supported in part by MDACC and NCI grant P30CA016672. K.M.M. was supported by an Ergon Foundation Postdoctoral Trainee Fellowship.
Author information
Authors and Affiliations
Contributions
R.K. conceptually designed the study, provided ideas for experiments and helped write the manuscript. V.S.L. helped with conceptual design of the study, provided ideas for experiments and executed the study. X.Z., V.S.L., E.S.F., J.K., K.M.M. and R.K. designed the experimental strategy and provided intellectual input. X.Z., V.S.L., E.S.F., J.K., J.D., H.S., T.M., L.M.B., O.V.P., E.L., C.E.S., S.I.P., A.K. and E.E. performed experiments and analyzed the data. B.L., C.C.W. and D.S. analyzed the sequencing and microarray data. X.Z., V.S.L., E.S.F., J.K. and B.L. helped write the manuscript and prepared the figures. J.A.K. provided intellectual input and insights into kidney metastasis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Cancer thanks Kai Kessenbrock, Vivek Mittal and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Lung and kidney fibrosis induction.
(a) Representative H&E staining, Sirius Red staining, and αSMA immunolabeling of the lung and kidneys of mice with and without fibrosis in the respective organs. Scale bar, 100 µm. (b) Quantification of positive Sirius Red and αSMA+ stained area in the indicated organs and groups, n = 5 to 6 mice per group. (c, d) Relative Fn1 and Col1a1 mRNA levels in the respective organs and groups, n = 3 mice per group. KF: kidney fibrosis; LF: lung fibrosis. (e) Representative images for GFP immunolabeling and quantification of percent of the GFP positive area in lung with or without lung fibrosis. Scale bar, 50 µm, n = 6 mice per group. (f) Representative images for GFP immunolabeling of tumor in the mice with or without lung fibrosis. n = 1 mouse per group. Scale bar, 25 µm. (g) Representative liver H&E images in 4T1 tumor orthotopic bearing mice after induction of lung fibrosis. Scale bar, 50 µm. Quantification of metastatic liver nodules in the indicated groups, n = 6 mice per group. (h) Representative images for GFP immunolabeling and quantification of percent of the GFP positive area in liver of mice with or without lung fibrosis. Scale bar, 50 µm, n = 6 mice per group. (i, j) Representative H&E images of heart (i) and intestine (j) in 4T1 tumor orthotopic bearing mice after induction of lung fibrosis. Scale bar, 50 µm. (k) Orthotopic 4T1 tumor volume over time in mice with or without kidney fibrosis, n = 6 mice per group. (l, m) Representative H&E images (l) of the lung of 4T1 orthotopic tumor bearing mice with or without kidney fibrosis and respective quantification (m) of metastatic area. Scale bar, 25 µm, n = 6 mice per group. (n) Representative images for GFP immunolabeling and quantification of percent of the GFP positive area in lung with or without kidney fibrosis. Scale bar, 50 µm, n = 6 mice per group. (o) Representative images for GFP immunolabeling for tumor in the mice with or without kidney fibrosis. Scale bar, 25 µm. (p) Representative liver H&E images in 4T1 tumor orthotopic bearing mice after induction of kidney fibrosis. Scale bar, 50 µm. Quantification of metastatic liver nodules in the indicated groups, n = 6 mice per group. (q) Representative images for GFP immunolabeling and quantification of percent of the GFP positive area in liver of mice with or without kidney fibrosis. Scale bar, 50 µm, n = 6 mice per group. (r) Representative H&E images of heart and intestine of 4T1 tumor orthotopic bearing mice after induction of kidney fibrosis. Scale bar, 50 µm. KF: kidney fibrosis; LF: lung fibrosis. Data are presented as mean values +/− SEM. For k: two-way ANOVA with Sidak’s multiple comparisons test. For all other panels: Unpaired two-tailed t test with Welch’s correction applied for unequal variances (determined by F-test). P values are listed, ns: not significant.
Extended Data Fig. 2 Fibrosis does not promote non-tropic metastasis.
(a) Representative H&E images of the kidneys of 4T1 orthotopic tumor bearing mice +/− lung or kidney fibrosis. Scale bar, 25 µm. (b) Number of mice with microscopic kidney tumors in the indicated groups (c) Representative images for GFP immunolabeling and quantification of percent of the GFP positive area in kidney +/− kidney fibrosis. Scale bar, 50 µm. (d) Number of mice with microscopic kidney tumors in the indicated groups. (e) Representative kidney H&E images in MMTV-PyMT transgenic mouse model with lung or kidney fibrosis. Scale bar, 25 µm. (f) Number of mice with microscopic kidney tumors in the indicated groups. (g) Representative immunofluorescence images of PyMT positive cancer area in tumor, lung, and kidney after kidney fibrosis in MMTV-PyMT transgenic mice. Scale bar, 25 µm. (h–j) Representative images and quantification of positive MTS and Sirius Red staining from the indicated groups. Scale bar, 25 µm, n = 6-7 mice per group. Data are presented as mean values +/− SEM. Unpaired two-tailed t test, with Welch’s correction applied for unequal variances (determined by F-test). P values are listed, ns: not significant.
Extended Data Fig. 3 Intracardiac cancer cell injection does not alter metastatic disease in renal fibrosis & Wound healing does not impact tropic and non-tropic site metastasis.
(a) Representative GFP and CD31 staining of the kidneys from mice with 4T1 orthotopic inoculation and 4T1 i.v. administration with or without kidney fibrosis. Scale bar, 25 µm. (b) Quantification of GFP+ cancer cells from lung after 4T1 i.v. administration for 10 days with or without perfusion by flow cytometry. n = 3 mice per group. Gating strategy shown in Supplementary Fig. 1A. (c) Quantification of GFP+ cancer cells from kidneys after 4T1 i.v. administration for 10 days with or without perfusion by flow cytometry, n = 3 mice per group. Gating strategy shown in Supplementary Fig. 1B. (d, e) QPCR quantification of GFP DNA (expressed 2-ΔCt after normalization to Nf1) from the lung (d) and kidney (e) of mice after 4T1 i.v. administration for 10 days with or without perfusion, n = 3 mice per group. (f) Representative H&E images and quantification of lung after intracardiac injection of 4T1 cancer cells together with induction of kidney fibrosis. Control, n = 5 mice; kidney fibrosis, n = 9 mice, Scale bar, 25 µm. (g) Representative H&E images and quantification of metastatic liver nodules after intracardiac injection of 4T1 cancer cells together with induction of kidney fibrosis. Control, n = 5 mice; kidney fibrosis, n = 9 mice. Scale bar, 50 µm. (h) Representative H&E images and number of mice with microscopic kidney tumors in the indicated groups after Intracardiac injection of 4T1 cancer cells together with induction of kidney fibrosis. Scale bar, 25 µm. (i) Representative images for GFP immunolabeling and number of mice with microscopic kidney tumors in the indicated groups after Intracardiac injection of 4T1 cancer cells together with induction of kidney fibrosis. Scale bar, 50 µm. (j) Orthotopic 4T1 tumor volume over time +/− cutaneous wound, n = 6 mice per group. (k) Representative H&E images of the lung of 4T1 orthotopic tumor bearing mice +/− cutaneous wound. Scale bar, 25 µm. (l) Quantification of metastatic area and surface lung nodules in 4T1 orthotopic tumor bearing mice with and without cutaneous wounds, n = 6 mice per group. (m) Tumor volume over time in MMTV-PyMT mice +/− cutaneous wound, n = 5 to 6 mice per group. (n) Representative lung H&E images from in MMTV-PyMT mice +/− cutaneous wound. Scale bar, 25 µm. (o) Quantification of metastatic area and surface lung nodules in MMTV-PyMT mice +/− cutaneous, n = 5 to 6 mice per group. (p) Wound closure rate in 4T1 orthotopic tumor bearing mice (n = 10 mice) and no tumor control (n = 4 mice). (q) Representative lung H&E images from mice with 4T1 i.v. injection and +/− cutaneous wound. Scale bar, 25 µm. (r) Quantification of metastatic area and surface lung nodules in mice with 4T1 i.v. injection +/− cutaneous wound, n = 5 mice per group. Data are presented as mean values +/− SEM or individual values shown. For j, m: two-way ANOVA with Sidak’s multiple comparisons test. For p: mixed effect model with Sidak’s multiple comparison test. For other panels: Unpaired two-tailed t test, with Welch’s correction applied for unequal variances (determined by F-test). P values are listed, ns: not significant.
Extended Data Fig. 4 Global gene expression profiling in fibrosis and serum cytokine analysis.
(a, b). Volcano plot and gene ontology (GO) enrichment ratio and false discovery rate (FDR) values associated with overlapped up-regulated and down-regulated genes. (c, d) GO enrichment ratio and FDR values in the lungs of mice with and without lung fibrosis (c) or with and without kidney fibrosis (d). (e, f) Overlapping up-regulated (e) or down regulated (f) genes in the lungs of healthy mice vs. mice with the lung fibrosis, and in the lungs of healthy mice vs mice with kidney fibrosis, with overlapped genes involved in vascular (f) and immune remodeling (e) pathways. (g) Cytokines profile of serum samples from healthy mice, mice with cutaneous wound, mice with lung fibrosis and kidney fibrosis, and heat map of the averaged relative cytokine levels in the indicated groups after normalization to healthy group (set to 1). Healthy, n = 2 mice; wound, n = 2 mice; lung fibrosis (LF), n = 3 mice; kidney fibrosis (KF), n = 3 mice. Cytokines showing minimal changes between healthy and wound (wound/healthy (W/H) ratio between 0.9 and 1.1) were then ranked based on levels in kidney fibrosis.
Extended Data Fig. 5 Immune cells infiltration in lung and kidney after fibrosis induction.
(a) Percentages of CD45+, CD3+, CD4+FoxP3− effector T (Teff) cells, CD8+, γδ+, CD4+FoxP3+ regulatory T (Treg) cells, NK1.1+ cells, CD11c+, CD19+, CD11b+, CD11b+Ly6C+Ly6G−and CD11b+Ly6G+ Ly6C− cells in lungs of mice with lung fibrosis or kidney fibrosis. Healthy lung, n = 6 mice; lung fibrosis (LF), n = 5 mice; healthy kidney, n = 6 mice; kidney fibrosis (KF), n = 5 mice. (b) Percentages of CD45+, CD3+, CD4+FoxP3− effector T (Teff) cells, CD8+, γδ+, CD4+FoxP3+ regulatory T (Treg) cells, NK1.1+ cells, CD11c+, CD19+, CD11b+, CD11b+Ly6C+Ly6G−and CD11b+Ly6G+Ly6C− cells in kidneys of mice with lung fibrosis or kidney fibrosis. Healthy lung, n = 5 mice; lung fibrosis (LF), n = 6 mice; healthy kidney, n = 6 mice; kidney fibrosis (KF), n = 4 mice. (c) Colocalization of FITC-dextran (green) and CD31 (red) in the liver of mice with and without after rAng2 treatment. Scale bar, 20 µm. Data are presented as mean values +/− SEM. Unpaired two-tailed t test, with Welch’s correction applied for unequal variances (determined by F-test). P values are listed, ns: not significant.
Extended Data Fig. 6 Recombinant Ang 2 enhances metastasis to the lung but not kidney.
(a) Representative lung H&E of mice treated with PBS (control) or recombinant Ang2 (rAng2). Scale bar, 25 µm. (b, c) Representative lung H&E of mice after induction of lung (b) or kidney fibrosis (c). Scale bar, 25 µm. (d) Quantification of edema area per whole lung area, n = 5 to 6 mice per group. (e) Quantification of pleural thickening of lung, n = 5 to 6 mice per group. (f) Representative kidney H&E images of mice treated with PBS (control) or rAng2. Scale bar, 25 µm. (g, h) Representative kidney H&E images of mice after induction of lung (g) or kidney fibrosis (h). Scale bar, 25 µm. (i) Representative lung H&E of mice treated with PBS (control) or recombinant Ang2 (rAng2) with 4T1 cells i.v. injection. Scale bar, 25 µm. (j) Representative kidney H&E of mice treated with PBS (control) or recombinant Ang2 (rAng2) with 4T1 cells i.v. injection. Scale bar, 25 µm. Data are presented as mean values +/− SEM. Unpaired two-tailed t test, with Welch’s correction applied for unequal variances (determined by F-test). P values are listed.
Extended Data Fig. 7 tSNE plots of Claudin genes in scRNA-seq data sets.
(a) tSNE plots of different Cldn genes in flow cytometry sorted CD31+ endothelial cells from mouse lung tissue34. (b) tSNE plots of different Cldn genes in endothelial cells from mouse lung tissue35. (c) tSNE plots of Claudin family genes in the endothelial cells from human lung scRNA-seq data35. (d) tSNE plots of Claudin family genes in the endothelial cells from mouse kidney scRNA-seq data36. (e) tSNE plots of Claudin family genes in the endothelial cells from human kidney scRNA-seq data37. Colors coded for the relative gene expressions.
Extended Data Fig. 8 Claudins expression in distinct cell types.
(a) Different Claudin expressions in different endothelial cell types of mouse lung1,34. (b, c) tSNE plots of Claudin 5 in different cell types of in mouse lung (b) and highly expressed in lung endothelial cells (c)35. (d) Different Claudins expression in different mouse lung cell types35. (e) Expression of diverse Claudins in different mouse kidney cell types36. (f, g) Diverse Claudins expression in different mouse34 (f) and human brain cell types (g)61. (h) Colocalization of Claudin 2 and CD31 in WT kidney cortex and medulla (individual images in Supplementary Fig. 5a, b). (i) Colocalization of Claudin 10 and CD31 in WT kidney cortex and medulla (individual images in Supplementary Fig. 5c, d). Scale bar, 50 µm (upper), 25 µm (lower). (j) Representative colocalization of Claudin 5 and CD31 in the lung of mice in the indicated groups. Scale bar, 20 µm. (k) Quantification of percentage of Claudin 5 and CD31 double positive area per visual field in the indicated groups, n = 5-9 mice per group. These images and graphs also shown in Supplementary Fig. 6. Data are presented as mean values +/− SEM. Unpaired two-tailed t test, with Welch’s correction applied for unequal variances (determined by F-test). P values are listed, ns: not significant.
Extended Data Fig. 9 Data Figure 9. Ang2 and Claudins signaling and Claudin2/10 suppression.
(a) Relative mRNA expression of Cldn5 in parental NIH-3T3 cells and NIH-3T3 cells treated with Adeno-CMV-luc (Ad-Ctrl) or Ad-CMV-CLDN5v2 (Ad-Claudin 5), n = 3 biologically independent experiments (with each 2 technical replicates). (b) Representative western blot and quantification of his-tagged Claudin 5 (His), Claudin 5, and β-actin in parental NIH-3T3 cells and NIH-3T3 cells treated with Adeno-CMV-luc (Ad-Ctrl) or Ad-CMV-CLDN5v2 (Ad-Claudin 5), n = 3 biologically independent experiments. Uncropped blots are shown in the Image Source Data File. (c) Relative mRNA expression of Cldn5 in NIH-3T3 cells with the indicated treatment. Dox on/off indicates doxycycline exposure, n = 4 biologically independent experiments (with each 2 technical replicates). (d) Orthotopic 4T1 tumor volume over time in the indicated groups, n = 5 mice per group. (e) CLDN5 mRNA levels in metastatic lungs of patients with breast cancer (n = 9) and healthy lungs (n = 6) (GSE193103)62. (f, g) Relative mRNA expression of Cldn2 (f) and Cldn10 (g) in kidney endothelial cells treated with the indicated guide RNAs, n = 3 biologically independent experiments (with each 2 technical replicates). (h) Relative mRNA expression of Cldn2 and Cldn10 in sorted E-cadherin+ epithelial cells and CD31+ endothelial cells, n = 3 mice per group. Gating strategy is shown in Supplementary Fig. 9. (i, j) Relative mRNA expression levels of Cldn2 and Cldn10 in sorted CD31+ endothelial cells (i) and E-cadherin+ epithelial cells (j) after treatment with the indicated guide RNAs, n = 3 mice per group. (k) Experimental design for the evaluation of the downregulation of Claudin 2 and Claudin 10 in the kidney of mice with 4T1 orthotopic tumors. (l) Orthotopic 4T1 tumor volume over time in the indicated groups. On day 14, LentiCRISPRv2 control vector, n = 5; LentiCRISPRv2 Cldn2 and Cldn10, n = 5; Day 18, LentiCRISPRv2 control vector, n = 5; LentiCRISPRv2 Cldn2 and Cldn10, n = 6; Day 22, LentiCRISPRv2 control vector, n = 8; LentiCRISPRv2 Cldn2 and Cldn10, n = 6 mice. (m) Representative H&E images of the lung of mice in the indicated groups. Scale bar, 25 µm. (n) Quantification of lung metastatic area. LentiCRISPRv2 control vector, n = 15; LentiCRISPRv2 Cldn2 and Cldn10, n = 20 mice. (o) Representative images for GFP immunolabeling and quantification of percent of the GFP positive area in lung. Scale bar, 50 µm, n = 9 mice per group. Data are presented as mean values +/− SEM, Min-to-Max in E. For a: one-way ANOVA test with Tukey’s multiple comparisons test. For d: two-way ANOVA with Sidak’s multiple comparisons test. For b, c, f, g: one-way ANOVA test with Dunnett’s multiple comparisons test. For e: two-tailed Mann-Whiney test. For i, j, n, o: Unpaired two-tailed t test, with Welch’s correction applied for unequal variances (determined by F-test).
Extended Data Fig. 10 Knockdown of Claudin 2/10 redirects metastasis to the kidney.
(a) Representative H&E images of the kidney of mice in the indicated groups. Scale bar, 50 µm (upper), 25 µm (lower). (b) Percentage of mice with kidney lesion in the indicated groups. LentiCRISPRv2 control vector, n = 15; LentiCRISPRv2 Cldn2 and Cldn10, n = 20 mice. (c) Representative images for GFP immunolabeling in kidney. Scale bar, 50 µm (upper), 25 µm (lower). (d) Number of mice with GFP positive kidney. (e) Quantification of percent of the GFP positive area in the indicated groups, n = 12 mice per group. (f) Orthotopic 4T1 tumor volume over time in the indicated groups. LentiCRISPRv2 control vector without fibrosis, n = 14; LentiCRISPRv2 control vector with fibrosis, n = 11; LentiCRISPRv2 Cldn2 and Cldn10 without fibrosis, n = 12 mice; LentiCRISPRv2 Cldn2 and Cldn10 with fibrosis, n = 10 mice. (g) Number of mice with GFP positive kidney in the indicated groups. LentiCRISPRv2 control vector without fibrosis, n = 14; LentiCRISPRv2 control vector with fibrosis, n = 11; LentiCRISPRv2 Cldn2 and Cldn10 without fibrosis, n = 12 mice; LentiCRISPRv2 Cldn2 and Cldn10 with fibrosis, n = 10 mice. (h) Representative GFP and CD31 staining of the kidneys in mice with Claudin 2 and Claudin 10 down regulation together with or without kidney fibrosis. (i) Representative GFP and CD31 staining in the metastatic kidney with Claudin 2 and Claudin 10 downregulation. For i-j, individual images are shown in Supplementary Fig. 10A-C. Scale bar, 25 µm. (j) Representative images for CD31 and quantification of percent of the CD31 positive area in tumor when downregulate Claudin 2/10 in the kidney. Scale bar, 50 µm. LentiCRISPRv2 control vector, n = 5; LentiCRISPRv2 Cldn2 and Cldn10, n = 7 mice. (k) Representative images for CD31 and quantification of percent of the CD31 positive area in tumor when downregulate Claudin 2 and Claudin 10 with or without kidney fibrosis. Scale bar, 50 µm. LentiCRISPRv2 control vector without fibrosis, n = 5; LentiCRISPRv2 control vector with fibrosis, n = 4; LentiCRISPRv2 Cldn2 and Cldn10 without fibrosis, n = 4 mice; LentiCRISPRv2 Cldn2 and Cldn10 with fibrosis, n = 6 mice. Data are presented as mean values +/− SEM. For b: two-sided Chi-square. For f: two-way ANOVA with Sidak’s multiple comparisons test. For j: Unpaired two-tailed t test, with Welch’s correction applied for unequal variances (determined by F-test). For k: one-way ANOVA with Tukey’s multiple comparisons test. P values are listed, ns: not significant.
Supplementary information
Supplementary Information
Supplementary Figs. 1–10, figure legends.
Supplementary Tables 1–6
Supplementary Tables 1–6.
Source data
Source Data
Uncropped western blots and cytokine array.
Source Data Fig. 1
Source data.
Source Data Fig. 2
Source data.
Source Data Fig. 3
Source data.
Source Data Fig. 4
Source data.
Source Data Fig. 5
Source data.
Source Data Fig. 6
Source data.
Source Data Fig. 7
Source data.
Source Data Fig. 8
Source data.
Source Data Extended Data Fig. 1
Source data.
Source Data Extended Data Fig. 2
Source data.
Source Data Extended Data Fig. 3
Source data.
Source Data Extended Data Fig. 4
Source data.
Source Data Extended Data Fig. 5
Source data.
Source Data Extended Data Fig. 6
Source data.
Source Data Extended Data Fig. 8
Source data.
Source Data Extended Data Fig. 9
Source data.
Source Data Extended Data Fig. 10
Source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhou, X., LeBleu, V.S., Fletcher-Sananikone, E. et al. Vascular heterogeneity of tight junction Claudins guides organotropic metastasis. Nat Cancer 5, 1371–1389 (2024). https://doi.org/10.1038/s43018-024-00813-1
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s43018-024-00813-1