Abstract
Peptide-based data storage offers a promising solution to the escalating digital data crisis. A key challenge is maintaining peptide stability over time after encoding information in amino acid sequences. Here, we present a simple and effective method to preserve peptides using a hydrogel composed of poly(N-isopropylacrylamide) and chitosan that forms a semi-interpenetrating polymer network. This hydrogel responds to temperature and pH stimuli to encapsulate and stabilize peptides from solution. Its thermoresponsive behavior enables efficient peptide concentration through repeated swelling and deswelling cycles. Strong electrostatic interactions between chitosan and peptides result in an ultrahigh data density of 2.44 × 1010 GB g−1. Accelerated aging tests under thermal and enzymatic conditions demonstrate significantly enhanced peptide stability, with original information fully recovered after 3.5 days at 70 °C, equivalent to over 600 years at 9.4 °C. This work establishes a durable, high-capacity platform for long-term peptide data storage.
Similar content being viewed by others
Introduction
The exponential growth of data, driven by the rapid evolution of information technology, has created significant challenges for data storage. Traditional storage media, such as hard drives and optical discs, are approaching their physical capacity limits, making them increasingly inadequate for the ever-growing demand for data storage1,2. In this context, biological information storage technologies, particularly DNA and peptide-based storage, have emerged as highly promising alternatives3,4,5,6.
DNA and peptides, as natural biopolymers, offer several unique advantages for information storage. DNA, with its high storage density and long-term stability, has been extensively studied for its potential to store vast amounts of data in a compact form7,8. Similarly, peptides, composed of amino acids linked by peptide bonds, provide a versatile platform for data encoding due to their structural diversity and ease of synthesis. The ability to encode information in the sequence of amino acids allows for high-density data storage, with the potential to store exabytes of data in just a few grams of material3,9. Moreover, peptides are inherently biodegradable and biocompatible, reducing the environmental impact compared to traditional storage media10.
Despite their advantages, peptide molecules are highly susceptible to degradation from environmental factors such as proteases, heat, moisture, and oxygen, which can lead to the loss of stored information11,12. Current preservation methods, including freezing, dehydration, and the use of salt ion buffer solutions, can extend the lifespan of peptides from several months to a few years. However, these methods are often costly, prone to contamination, and offer limited protection periods13,14. As a result, the development of more robust and long-term preservation strategies for peptide-based information storage remains a critical area of research. Recent advancements in peptide preservation are still in its early stages, with relatively few reports. For instance, Zheng et al.15 proposed an innovative approach involving the fusion of mirror-image proteins onto plastic films, which significantly improved the stability of stored information. However, challenges such as long-term, high-density preservation and flexible, secure access to the stored data remain unresolved. These limitations highlight the need for advanced preservation techniques that can ensure the integrity of peptide-based information over extended periods.
Hydrogels, with their three-dimensional network structure, have emerged as a promising medium for the long-term preservation of peptide-based information16,17,18. The unique properties of hydrogels, such as their high-water absorption capacity and ability to minimize direct contact between peptides and harmful environmental factors, make them ideal for protecting peptides from degradation19,20. Additionally, stimuli-responsive hydrogels, which can respond to environmental changes such as temperature, pH, light, or specific chemicals, offer the potential for controlled release and retrieval of stored information21,22. By embedding peptide-based information storage media within such hydrogels, it is possible to achieve both high-density storage and long-term preservation, while also enabling flexible and secure access to the data23.
Among the various types of hydrogels, PNIPAM (Poly(N-isopropylacrylamide)) hydrogels have garnered significant attention due to their unique thermos-responsive properties. PNIPAM hydrogels exhibit a lower critical solution temperature (LCST) and volume phase transition temperature (VPTT) of ~32 °C, below which they remain fully swollen. Above this temperature, the polymer network transitions from hydrophilic to hydrophobic, causing the hydrogel to release its absorbed water24,25,26. This property, combined with the ability to chemically modify PNIPAM through copolymerization or the formation of semi-interpenetrating polymer networks (semi-IPN), enhances its functionality and makes it suitable for multimodal applications27,28. Chitosan (CS), a natural polysaccharide with a pKa value of around 6.5, can be incorporated into PNIPAM hydrogels to improve their affinity for negatively charged peptides29,30. At pH values below 6.5, CS carries a positive charge, allowing it to bind electrostatically with negatively charged peptides31,32. This interaction not only increases the peptide loading capacity of the hydrogel but also provides a mechanism for controlled peptide release by adjusting the pH33,34. Importantly, the introduction of CS does not affect the LCST of PNIPAM, enabling peptide storage and retrieval at lower temperatures35. Furthermore, CS has been shown to protect negatively charged proteins, suggesting its potential as a protective agent for peptides within hydrogels36,37.
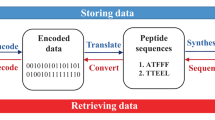
Here, we investigate PNIPAM/CS semi-IPN hydrogels for peptide-based information storage (Fig. 1). Textual data is encoded into peptide sequences, which are absorbed by hydrogel at 20 °C under acidic conditions. Swelling and dehydration cycles at 45 °C anchor peptides via electrostatic interactions with protonated CS, enabling high-density storage. For retrieval, the hydrogel is immersed in an alkaline buffer, where CS deprotonates, weakening peptide binding and allowing efficient extraction. Optimized pH conditions, storage density calculations, and accelerated aging tests confirm the hydrogel’s protective performance38. Mass spectrometry successfully decodes peptides back into original information, demonstrating the system’s feasibility. This approach combines the stability and flexibility of hydrogels with chitosan’s protective properties, offering a promising solution for high-density, long-term data storage, addressing modern data storage challenges.
Created in BioRender. C, X. (2025) https://BioRender.com/y1pe53r.
Results and discussion
Peptide encoding and decoding
To fully harness the potential of peptide-based storage technology for big data applications, we strategically selected eight amino acids for data encoding. A key advantage of this selection is that peptides composed of these amino acids exhibit an isoelectric point of ~3.5. This property ensures that the peptides carry a negative charge in environments with a pH above 3.5, facilitating strong electrostatic interactions with positively charged hydrogel networks. Furthermore, each amino acid encodes 3 bits of data, significantly enhancing information storage density. While increasing the variety of amino acids could theoretically boost storage capacity, we balanced this consideration with the practical challenges of synthesis and sequencing costs. This approach optimizes both storage efficiency and experimental feasibility. Using our tailored encoding rules, we successfully encoded the text “Welcome to Shanghai University” into 5 peptides comprising 85 amino acids, demonstrating the system’s effectiveness. (Supplementary Fig. S3).
Preparation and characterization of PNIPAM/CS semi-IPN hydrogels
We synthesized a series of PNIPAM/CS semi-IPN hydrogels with varying CS concentrations to achieve high peptide loading capacity, stable preservation, and precise control over peptide release via temperature and pH adjustments. As shown in Fig. 2a, the IPN CS3 hydrogel beads exhibit diameters of ~2 mm when dry and 5 mm when fully swollen. Notably, unlike pure PNIPAM hydrogel, which turns transparent upon swelling at 25 °C39, the PNIPAM/CS semi-IPN hydrogel retains a white appearance in both dry and swollen states (see Supplementary Fig. S4 for details).
a Fully dried and fully expanded states of IPN CS3 hydrogel. The scale bar represents 10 mm. b DSC thermograms showing VPTT of pure PNIPAM and PNIPAM/CS semi-IPN hydrogels with varying CS content. c Equilibrium swelling ratio of hydrogels measured after 2 days in DI water at 20 °C and 1 h at 45 °C. d SEM images of hydrogel cross-sections. The scale bar represents 50 μm. e, f Swelling kinetics of hydrogels at 20 °C and deswelling kinetics at 45 °C. g A complete suction and discharge water cycle: swelling at 20 °C and deswelling at 45 °C. Error bars represent mean ± standard deviation (SD).
To investigate the thermoresponsive behavior of the PNIPAM/CS semi-IPN hydrogels, we measured the VPTT using differential scanning calorimetry (DSC). As depicted in Fig. 2b, the VPTT of pure PNIPAM is 30.24 °C, slightly below its typical LCST of 32 °C. For semi-IPN hydrogels with increasing CS content (IPN CS1, IPN CS2, and IPN CS3), the VPTTs are 30.92 °C, 29.57 °C, and 29.26 °C, respectively. This minimal variation suggests that the CS network, acting as a secondary structure without reacting with NIPAM (N-Isopropylacrylamide), preserves the thermoresponsive properties of PNIPAM28,35. The equilibrium swelling ratio (ESR) further characterizes the hydrogels’ thermoresponsive behavior. As illustrated in Fig. 2c, IPN CS1, IPN CS2, and IPN CS3 exhibit reduced swelling ratios below their respective VPTTs, attributed to the semi-IPN structure limiting hydrogel microsphere swelling40. Higher CS content results in smaller and more numerous pores (Fig. 2d), further decreasing the swelling ratio41. Figure 2e, f demonstrate the hydrogels’ swelling and deswelling kinetics at 20 °C and 45 °C, respectively. At 20 °C, the hydrogels absorb significant water within 4–6 h, while at 45 °C, they rapidly release most absorbed water in under 10 min. Notably, the PNIPAM-only hydrogel exhibits a higher residual swelling ratio compared to semi-IPN samples at 45 °C (Fig. 2f). This is likely due to the formation of a dense skin layer on the hydrogel surface upon collapse, which impedes water diffusion from the core42. In contrast, the incorporation of hydrophilic linear CS chains in the semi-IPN structure facilitates internal water transport, resulting in more efficient dehydration and a lower final swelling ratio43. Based on these findings, we established optimal operational parameters: 4 h of peptide absorption at 20 °C and 10 min of release at 45 °C, as summarized in Fig. 2g.
High peptide loading capacity and peptide recovery of PNIPAM/CS semi-IPN hydrogels
Figure 3a illustrates the peptide absorption/desorption mechanism of the PNIPAM/CS semi-IPN hydrogel. To enable precise peptide quantification, we established a standard peptide concentration curve (Supplementary Fig. S5). We first determined the optimal pH for peptide absorption and desorption, as shown in Fig. 3b, c. The IPN CS3 hydrogel achieved maximum peptide loading at pH 5, where electrostatic interactions between peptides and the hydrogel are strongest. For peptide release, an elution buffer with pH 9.5 proved most effective, while incomplete release occurred between pH 7 and 8.5. This behavior is linked to chitosan’s pKa (~6.5), requiring a higher pH for complete deprotonation and efficient peptide desorption44. Consequently, we selected pH 5 for absorption and pH 9.5 for release to optimize peptide loading and desorption. Zeta potential measurements at these pH values (Fig. 3d) confirmed that peptides and the hydrogel carry opposite charges at pH 5, while at pH 9.5, chitosan’s deprotonation minimizes electrostatic interactions, facilitating peptide release.
a Schematic of peptide absorption in PNIPAM/CS semi-IPN hydrogel via swelling/deswelling in peptide solution, followed by release using elution buffer (pH >9.5). Created in BioRender. C, X. (2025) https://BioRender.com/l43dk1kb Effect of initial solution pH on peptide absorption. Note: sum of concentration reflects peptide decrease in the initial absorption solution. c Effect of elution buffer pH on the peptide desorption. Note: sum of concentration reflects peptide concentration in released buffer from IPN CS3 hydrogel after deswelling. d Zeta potential analysis of peptides and hydrogels in buffers at pH 5 and 9.5. e Peptide loading capacity in hydrogel, reflected by decrease in peptide concentration in the initial absorption solution after multiple absorption cycles. f Change in peptide concentration in released solution after deswelling at 45 °C in multiple cycles. g Comparison of peptide loading efficiency between dynamic swelling/deswelling cycles and static immersion. “ns” indicates no significant difference; “*” (p < 0.05) and “**” (p < 0.01) indicate statistically significant differences. Error bars represent mean ± standard deviation (SD).
We also examined the impact of CS content on peptide loading capacity. As shown in Fig. 3e, the IPN CS3 hydrogel exhibited the highest loading capacity, while pure PNIPAM had the lowest, indicating that higher CS content enhances peptide loading. This is attributed to the increased zeta potential of the hydrogel with higher CS content (Fig. 3d), reflecting a stronger positive surface charge and enhanced electrostatic attraction to negatively charged peptides45. Once the hydrogel reaches saturation, peptides can be efficiently released using a pH > 9.5 elution buffer (Fig. 3f). At high pH, deprotonation of chitosan’s amine groups disrupts CS-peptide complexes, enabling rapid peptide release.
To examine whether dynamic swelling and deswelling cycles improve adsorption efficiency, we compared two loading strategies: (1) four cycles of thermal swelling/deswelling and (2) static immersion for an equivalent total duration (16 h 40 min). As shown in Fig. 3g, cyclic loading significantly increased peptide uptake. This improvement likely stems from two synergistic effects: repeated swelling disrupts local equilibrium and promotes continuous diffusion into the hydrogel matrix, while deswelling compresses the network and enhances peptide retention via electrostatic attraction with CS chains.
Taken together, these results demonstrate that the PNIPAM/CS semi-IPN hydrogel enables rapid, efficient, and reversible peptide loading and release. Its dynamic adsorption behavior, combined with favorable thermoresponsive properties, makes it a promising candidate for high-efficiency bioinformation storage.
Figure 4a shows the swelling ratio changes of the IPN CS3 hydrogel over time during multiple peptide absorption cycles. As depicted in Fig. 4b, the time required for the IPN CS3 hydrogel to fully absorb a constant volume (0.034 mL) of peptide solution increases from 4 h to 5.27 h, while the deswelling time at 45 °C remains constant at 10 min. A similar trend was observed by Fei et al.23 in TRFG hydrogels, who attributed this behavior to the formation of complexes between protonated amine groups of HPEI (polyethyleneimine) and negatively charged DNA molecules, reducing the availability of protonated functional groups due to charge compensation. In contrast, Wu et al.46 suggested that hydrogel reswelling kinetics are slower than initial swelling kinetics. To clarify this, we measured the swelling time of the IPN CS3 hydrogel in DI water and found no increase in swelling duration (Supplementary Fig. S6), supporting Fei et al.’s explanation23. This indicates that PNIPAM/CS semi-IPN hydrogels are reusable.
a Swelling ratio changes of IPN CS3 hydrogel over time for different peptide absorption cycles. b Swelling time required for complete absorption of 0.034 mL initial peptide solution (pH 5) in different absorption cycles. c Maximum peptide loading capacity of IPN CS3 hydrogel in initial peptide solutions of varying concentrations. Note: sum of concentration reflects peptide decrease in the initial absorption solution. Different letters above the bars indicate statistically significant differences. (one-way ANOVA, LSD test, p < 0.05). d Data storage density of peptide-PNIPAM/CS semi-IPN hydrogel systems, with light blue background indicating DNA encapsulation system for comparison. e Impact of competitive charged species (BSA, HS, and NaCl) on peptide loading (light pink background) and stability (light blue background) in IPN CS3 hydrogels. stability was assessed after 7-day exposure under simulated ambient conditions (30 °C, 50% humidity, 21% oxygen, no light shielding). Note: sum of concentration reflects peptide concentration in released buffer from IPN CS3 hydrogel after deswelling. Error bars represent mean ± standard deviation (SD).
To determine the minimum peptide concentration required for saturation, we evaluated the peptide loading capacity of the IPN CS3 hydrogel after 10 absorption cycles in solutions of varying concentrations. As shown in Fig. 4c, the hydrogel did not reach saturation at peptide concentrations of 100 and 200 μg mL−1 but achieved saturation at 300 μg mL−1 or higher, indicating that 300 μg mL−1 is the threshold concentration for saturation.
We then calculated the information storage density of the peptide-PNIPAM/CS semi-IPN hydrogel system (Fig. 4d). For the IPN CS3 hydrogel, the peptide concentration in the elution solution (0.3 mL) was 107 μg mL−1, and the weight of a single hydrogel bead was 0.0023 g (Supplementary Fig. S7), yielding a peptide loading density of 0.014 g g−1. Given the average weight of the eight amino acids (≈2.0 × 10−22 g), 1 g of IPN CS3 hydrogel contains ~7 × 10¹⁹ amino acids. With each amino acid encoding 3 bits of data, the storage capacity of 1 g of hydrogel is ~2.1 × 10²⁰ bits, equivalent to 2.44 × 10¹⁰ GB g−1. This density is roughly three times higher than that of the current best DNA-based encapsulation technology (Layer-by-Layer)47. These results highlight the significant competitive advantage of the peptide-PNIPAM/CS semi-IPN hydrogel system in terms of information storage density.
To further evaluate the practical robustness of this system, we next examined how common charged species may influence peptide loading and stability in the IPN CS3 hydrogel (Fig. 4e). We introduced bovine serum albumin (BSA), heparin sodium (HS), and sodium chloride (NaCl) into the system. BSA is a negatively charged, amphiphilic protein with multiple binding sites48, while HS is a highly sulfated polysaccharide bearing dense negative charges and is commonly used as a model polyanion49. The peptide concentration was maintained at 1 mg mL−1 in all conditions. In the presence of 0.9% NaCl, peptide uptake into the IPN CS3 hydrogel was reduced to 16.5 μg mL−1, suggesting that ionic shielding from high concentrations of monovalent ions (Na⁺ and Cl⁻) effectively disrupted electrostatic interactions between the peptide and the hydrogel network, thereby preventing retention50. In contrast, the addition of BSA or HS (each at 1 mg mL−1) led to peptide loadings of 84.2 and 50.0 μg mL−1, respectively. These results indicate that although both species compete for electrostatic binding sites within the hydrogel, the degree of interference differs. Interestingly, after a 7-day exposure under simulated ambient conditions (30 °C, 50% humidity, 21% oxygen, no light shielding), the presence of these charged species did not significantly affect the relative stability of the encapsulated peptide. This suggests that once encapsulated, peptides remain well protected against environmental degradation, even in the presence of other charged molecules.
These results demonstrate that certain small charged molecules can compete with peptides for binding to the hydrogel, leading to reduced loading efficiency. Fortunately, they did not compromise the stability of the stored peptides. To mitigate this issue, peptide-loaded hydrogels can be stored in centrifuge tubes to minimize contact with interfering substances. This simple, low-cost approach provides an effective way to maintain peptide loading capacity and preserve data integrity.
Long-term preservation and data recovery of peptides in PNIPAM/CS semi-IPN hydrogels
We further evaluated the long-term protection and storage stability of peptides encapsulated within the IPN CS3 hydrogel by subjecting both free and encapsulated peptides to enzymatic and high-temperature accelerated aging tests (Fig. 5a, b). As shown in Fig. 5a, the enzymatic degradation rates of the five peptides varied, with peptide b exhibiting the slowest degradation. Encapsulated peptides demonstrated significantly reduced degradation rates compared to free peptides. After 12 h of enzymatic treatment, free peptides (excluding peptide b) retained less than 1% of their original mass, while encapsulated peptides retained 16% to 42%. For peptide b, the residual amounts were 18% for free peptides and 46% for encapsulated peptides. Similarly, Fig. 5b illustrates the thermal degradation kinetics of mixed peptides at 60 °C, 65 °C, and 70 °C. Encapsulated peptides exhibited markedly slower thermal degradation, retaining 12% to 55% of their original mass after 14 days, compared to only 1% to 3% for free peptides. These results confirm that the IPN CS3 hydrogel effectively shields peptides from both enzymatic and thermal degradation.
a Enzymatic hydrolysis kinetics of pure peptide powder and peptides encapsulated in PNIPAM/CS semi-IPN hydrogels, with 5 peptide codes encoding “Welcome to Shanghai University”. b Thermal degradation kinetics of pure peptide powder and peptides encapsulated in PNIPAM/CS semi-IPN hydrogels. c Remaining peptide quantity in PNIPAM/CS semi-IPN hydrogels with varying CS content after 12 h of enzymatic hydrolysis. Different letters above the bars indicate statistically significant differences. (one-way ANOVA, LSD test, p < 0.05). d Remaining peptide quantity in PNIPAM/CS semi-IPN hydrogels with varying CS content after 3.5 d at 70 °C. Different letters above the bars indicate statistically significant differences. (one-way ANOVA, LSD test, p < 0.05). e Half-life of peptide stored in IPN CS3 hydrogel extrapolated according to the Arrhenius Equation. f Experimental and modeled degradation curves of peptides encapsulated in IPN CS3 hydrogels under environmental exposure (30 °C, 50% humidity, 21% oxygen, no light shielding) over 0, 7, 15, and 30 days. The modeled curve was generated based on the Arrhenius equation and a first-order kinetic model. g Correlation analysis between experimental and modeled peptide degradation curves. Ce0 and Cm0 denote peptide concentrations at day 0 for the experimental and modeled curves, respectively, with the same notation for other time points. Error bars represent mean ± standard deviation (SD).
To maximize the protective properties of the hydrogel for peptides, we aim to leverage the enhanced protective capabilities provided by CS within the hydrogel matrix. By comparing the protective performance of PNIPAM/CS semi-IPN hydrogels with varying CS content (Fig. 5c, d), we found that pure PNIPAM hydrogel, lacking CS, offered the least protection. Increasing CS content significantly improved protective performance, peaking at a relative CS content of approximately 2. This aligns with findings by Masuoka et al.37, who reported that CS protects fibroblast growth factor-2 (FGF-2) from thermal and proteolytic degradation through complex formation involving electrostatic interactions and hydrogen bonding. Collado-González et al.36 further demonstrated that CS stabilizes negatively charged protein nanoparticles via molecular entanglement and bridging. These mechanisms support our hypothesis that positively charged CS provides additional protection for encapsulated peptides.
Using a first-order kinetic model and the Arrhenius equation38, we calculated the activation energy for peptide decay in the IPN CS3 hydrogel as 145.9 kJ mol−1 (Supplementary Fig. S8 and Fig. S9). Building on the Accelerated Stability Assessment Program (ASAP) applied by Waterman et al.51 to evaluate peptide stability, we simulated the long-term decay behavior of encapsulated peptides. The results, shown in Fig. 5e, reveal a strong temperature dependence on peptide half-life. At 20 °C, encapsulated peptides have a half-life of approximately 62 years (green triangle), extending to about 600 years at 9.4 °C (blue circle). While this does not match the >500-year stability reported for some systems at room temperature52,53, it surpasses the half-life of DNA encapsulated in silica at 9.4 °C (~500 years) reported by Grass et al.38.
To further evaluate the long-term applicability of our storage system under practical conditions, we conducted additional environmental exposure experiments simulating realistic conditions (30 °C, 50% relative humidity, 21% oxygen, and no light shielding). Peptide concentrations were monitored at 0, 7, 15, and 30 days. The experimental degradation curve of the encapsulated peptides closely matched the modeled degradation curve predicted by the Arrhenius equation and first-order kinetics (Fig. 5f). Correlation analysis (Fig. 5g) between the two curves yielded an R² of 0.988 and a slope (k) of 1.080, indicating a high level of consistency with a slight systematic deviation. These findings underscore the exceptional potential of our peptide-PNIPAM/CS semi-IPN hydrogel system for long-term information storage.
To demonstrate the accessibility of information stored in synthetic peptides after prolonged thermal treatment, peptides encapsulated in the IPN CS3 hydrogel and aged at 70 °C for 3.5 days (degraded by one half-life) were separated using liquid chromatography (LC) and fragmented to generate MS/MS (tandem mass spectrometry) spectra. This process allowed for the reconstruction of amino acid sequences based on the mass differences between fragment ions (Fig. 6a–f). Previous studies by Ng et al.3 and Zhang and Zhu54 have successfully applied this method to sequence information-storing peptides. Using the predefined mapping rules (Supplementary Fig. S1), the obtained peptide sequences were converted into binary codes and subsequently decoded into the original data. The results confirmed that the sequences of all five peptides were accurately retrieved, enabling the complete reconstruction of the original message (Fig. 6g): “Welcome to Shanghai University”. By successfully recovering data from peptides degraded by one half-life and considering the stability data presented in Fig. 5e, we conclude that digital information can be stably preserved in the IPN CS3 hydrogel for up to 600 years under conditions such as those in Zurich (9.4 °C). This highlights the system’s potential for ultra-long-term data storage.
Limitations and future directions of peptide-based data storage
Selective retrieval of specific peptide sequences remains challenging in peptide-based molecular storage. Our current system uses bulk pH-triggered release, limiting addressability. To address this, we propose using flow cytometry to sort hydrogel microspheres by size or fluorescence, enabling high-throughput, multiplexed, and non-destructive retrieval of specific data fragments55.
On the decoding side, MS/MS has demonstrated effectiveness for short peptide sequencing but faces scalability limitations. These stem from its reliance on predefined peptide databases, relatively low throughput, and the inability to amplify peptides like DNA via PCR (Polymerase Chain Reaction), which hampers the detection of low-abundance sequences. These factors collectively constrain the scalability and sensitivity of peptide decoding. Nanopore peptide sequencing offers a promising alternative. It enables direct, single-molecule, amplification-free decoding with high sensitivity and single–amino acid resolution. Unlike MS/MS, nanopore approaches could provide real-time, database-independent access to peptide information, potentially unlocking the scalability needed for large peptide data archives56,57,58.
Compared to the more mature DNA data storage systems, peptide-based storage currently faces challenges in synthesis cost and read/write throughput. However, peptides offer significant advantages in terms of high storage density, long-term stability, and flexible chemical encoding. With ongoing advancements in peptide synthesis and emerging sequencing technologies such as nanopore peptide sequencing, these limitations are expected to be overcome. Consequently, peptide-based storage holds great potential to become a valuable complement in the molecular data storage landscape, enabling more efficient and scalable information storage solutions in the future.
Conclusion
We present a groundbreaking and efficient long-term preservation strategy for peptide-based information storage media, leveraging a thermally and pH-responsive PNIPAM/CS semi-IPN hydrogel to encapsulate and protect peptides. This innovative approach achieves an ultrahigh information density of 2.44 × 10¹⁰ GB g−1, surpassing the current best DNA encapsulation technology (Layer-by-Layer) by a factor of three. Through accelerated aging experiments under high-temperature and enzymatic conditions, we demonstrate that the PNIPAM/CS semi-IPN hydrogel significantly enhances peptide stability, offering robust protection against degradation. Remarkably, we successfully recovered the original information from peptides stored in the hydrogel after accelerated aging at 70 °C for 3.5 days, equivalent to a stable preservation period of 600 years at 9.4 °C. These results underscore the exceptional potential of PNIPAM/CS semi-IPN hydrogels as a durable and high-capacity platform for long-term peptide-based information storage. Our findings represent a transformative advancement toward developing a peptide-based storage system that could outperform conventional silicon-based materials, paving the way for next-generation data storage solutions.
Methods
Materials
NIPAM and CS (with a deacetylation degree of 90% and an average molecular weight of 2.0 × 10⁵) were purchased from Shanghai Maclin Biochemical Technology Co., Ltd. (China). Both NIPAM and CS were purified prior to use, and the detailed purification procedures are provided in Supplementary Note S1. Potassium persulfate (KPS), N, N, N′, N′-tetramethyl ethylenediamine (TEMED), N, N′-methylenebisacrylamide (MBA), acetic acid (HAc), bovine serum albumin (BSA), heparin sodium (HS) and sodium hydroxide (NaOH) were of analytical grade and also obtained from Shanghai Maclin Biochemical Technology Co., Ltd. (China). The peptide concentration was determined using the Pierce Quantitative Fluorometric Peptide Assay Kit (Thermo Fisher Scientific, USA).
Peptides coding and synthesis
The encoding and decoding methods are illustrated in Supplementary Fig. S1, and the peptide design is detailed in Supplementary Note S2. To facilitate the conversion between peptide codes and digital information, a custom program was developed using R, as shown in Supplementary Fig. S2 and Notes S3 and S4.
The peptides were synthesized by Synpeptide Co., Ltd. (China) via the solid-phase peptide synthesis method. After synthesis, the peptides were solubilized in a 0.1 M NaHCO₃ solution. The pH of the solution was adjusted as needed using either a 1% (v: v) HAc solution or a 0.1 M NaOH solution.
Synthesis of Semi-IPN hydrogels
PNIPAM/CS semi-IPN hydrogels were synthesized via free radical polymerization, using MBA as the cross-linker and KPS/TEMED as the redox-initiator pair at 20 °C59. Briefly, NIPAM was dissolved in a 1% (v: v) HAc solution containing varying amounts of CS in a three-necked round-bottom flask. The mixture was deoxygenated by purging with nitrogen for 20 min and then cooled to 4 °C for several hours. Under continuous nitrogen flow and at low temperature, freshly prepared TEMED (5 vol%) and KPS (10 wt%) solutions, pre-cooled to 4 °C, were sequentially added to the mixture with continuous stirring for 20 s. The resulting mixture was quickly transferred into a mold and placed in a nitrogen-purged desiccator for low-temperature polymerization at 20 °C for 24 h, yielding small spherical hydrogels with an approximate diameter of 3 mm. The semi-IPN hydrogels were thoroughly washed by immersion in DI water for 3 days, with periodic water change, and subsequently dried at 60 °C. The sample codes and feed composition are summarized in Table 1.
Characterization of Semi-IPN hydrogels
The interior morphology of the lyophilized fully swollen semi-IPN hydrogels was characterized using SEM (Thermo Scientific Quattro ESEM, USA). Before the SEM observation, the samples were sputter-coated with platinum to improve their conductivity.
The VPTT of both the pure PNIPAM hydrogel and the semi-IPN hydrogels was determined by DSC (DSC TA570, TA Instruments). Fully swollen hydrogel samples (~10 mg) were sealed in aluminum hermetic pans, equilibrated at 10 °C, and then heated to 60 °C at a rate of 2 °C min−1. The swelling ratio (SR) and equilibrium swelling ratio (ESR) of the hydrogels in DI water were measured using a gravimetric method. The SR and ESR were calculated using the following equations:
where wt is the wet weight of the hydrogel, wd is the dry weight of the hydrogel, and we represents the weight of the hydrogel at equilibrium.
The zeta potential of the hydrogels was measured by preparing a suspension of pulverized hydrogel and analyzing it using a Particle Size and Zeta Potential Analyzer (Zeta Sizer 3000 HSA, UK). Similarly, the zeta potential of the peptides was determined by analyzing a peptide solution with the same instrument.
Peptide capacity and peptide recovery of hydrogels
The peptide concentration was determined using a standard curve generated from a series of peptide solution dilutions (0, 3.125, 6.25, 12.5, 25, 50, 100 μg mL−1). Subsequently, the synthesized hydrogels were used to absorb 0.3 mL of the initial peptide solution at 20 °C. The temperature of the hydrogels beads was then increased to 45 °C in an incubator for ~10 min to release the absorbed water. The peptide concentrations in both the initial solution and the released water were quantified using a multifunctional microplate reader (SpectraMax iD3, USA). Absorption cycles were repeated until no significant changes in peptide concentrations were observed in the original solution. To facilitate peptide release, the peptide-loaded hydrogels were immersed in 0.3 ml of elution buffer with pH values ranging from 7 to 10. By alternating the temperature between 20° and 45 °C, the elution buffer was absorbed and released multiple times to ensure complete peptide recovery. The peptide concentration in the elution buffer released from the hydrogels was then determined using the multifunctional microplate reader.
Accelerated aging of peptides
Enzymatic accelerated aging: The enzymatic accelerated aging of peptides was performed as follows: A peptide solution with a concentration of 2 mg mL−1 was prepared and mixed with human serum at a 4:1 ratio (v: v). The mixture was incubated at 37 °C for varying durations (0, 1, 2, 4, 8, and 12 h). At each time point, 50 μL of ethanol was added to 50 μL of the mixture to terminate the reaction. The mixture was centrifuged at 10,000 rpm for 10 min, and the supernatant was collected for high-performance liquid chromatography (HPLC) analysis at a detection wavelength of 220 nm. The remaining peptide amount was quantified by comparing the peak areas at each time point to that at 0 h.
For peptides encapsulated within hydrogels, a solution of 400 μL human serum and 100 μL DI water was prepared. Then, 200 μL of this solution was mixed with the hydrogel and incubated for the specified durations. After incubation, the serum was removed, and the hydrogel was rinsed twice with DI water. The released peptides were analyzed using HPLC.
Thermal accelerated aging: Thermal accelerated aging experiments were conducted based on the methodologies reported by Grass et al.38. Accelerated aging tests were performed in ovens set at three temperatures: 60 °C, 65 °C, and 70 °C. Freeze-dried peptide powders and peptide-loaded hydrogels were placed in open Eppendorf tubes, which were then sealed within glass containers. To simulate typical indoor storage conditions, all experiments were conducted in the presence of 21% oxygen and without light shielding. Additionally, a saturated NaBr solution was placed in each container to maintain a relative humidity of approximately 50%.
Kinetic analysis: The first-order rate constant (k) for the degradation of peptides encapsulated within hydrogels was determined at the three temperatures using the exponential decay formula:
Where, C is the peptide concentration at time t, C0 is the initial peptide concentration, k is the first-order rate constant, and e is Euler’s number (~2.71828).
The rate constants were further analyzed using the Arrhenius equation to calculate the activation energy (Ea) for peptide degradation:
Where A is the pre-exponential factor, R is the ideal gas constant (8.314 J (mol K)−1), T is the absolute temperature in Kelvin, and e is Euler’s number.
Using the activation energy (Ea) and the pre-exponential factor (A), the half-life of the peptides encapsulated in the hydrogels was calculated for various temperatures by integrating Eqs. (3) and (4).
sequencing analysis
The peptide mixtures were separated using a Thermo Vanquish UPLC system equipped with a C18 column (Waters ACQUITY UPLC Peptide BEH C18 Column, 2.1 × 100 mm, 1.7 μm particle size, 130 Å pore size). The mobile phases consisted of (A) 0.1% formic acid in water and (B) 0.1% formic acid in acetonitrile. The flow rate was set to 0.35 mL min−1, and the column temperature was maintained at 40 °C. The gradient program was as follows: 5% B from 0 to 1 min, a linear increase from 5% B to 95% B from 1 to 12 min, and 95% B held for 1 min.
MS/MS analysis was performed using Q-Exactive Plus Orbitrap mass spectrometer (Thermo Fisher Scientific, San Jose, CA) in positive ion mode. The electrospray ionization (ESI) parameters were set as follows: spray voltage at +3800 V, capillary temperature at 320 °C, and auxiliary gas heater temperature at 300 °C. Data-dependent acquisition (DDA) mode was used for MS/MS data collection. The MS1 scan range was set to m/z 400–2000 Da with a resolution of 70,000, and MS/MS spectra were acquired with a minimum mass of 70 Da and a resolution of 17,500. High-energy collision dissociation (HCD) at 20 eV was employed for peptide fragmentation.
The MS/MS spectra were analyzed for sequence identification using Thermo Proteome Discoverer 2.5 software. A custom FASTA file was used as the search database. The error thresholds were set to <10 ppm for precursor ions and <0.2 Da for fragment ions, with no enzyme digestion specified. Peptide-spectrum matches (PSM) were further processed and visualized using Origin software.
Reproducibility and data presentation
All experiments were performed with at least three independent replicates. Data are presented as mean ± standard deviation (SD). To verify reproducibility, key experiments were repeated using at least two separate batches of hydrogels or peptides, which showed consistent results across batches.
To improve figure clarity, error bars were omitted in Figs. 2e, f, and 3b, c, e, f. Corresponding SD values are available in the Supplementary Tables S1–S3.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Evans, R. F. L., Chantrell, R. W., Nowak, U., Lyberatos, A. & Richter, H. J. Thermally induced error: density limit for magnetic data storage. Appl. Phys. Lett. 100, 102402 (2012).
Charap, S. H., Lu, P. L. & He, Y. Thermal stability of recorded information at high densities. IEEE Trans. Magn. 33, 978–983 (1997).
Ng, C. C. A. et al. Data storage using peptide sequences. Nat. Commun. 12, 4242 (2021).
Mao, C. et al. Metal–organic frameworks in microfluidics enable fast encapsulation/extraction of DNA for automated and integrated data storage. ACS nano 17, 2840–2850 (2023).
Church, G. M., Gao, Y. & Kosuri, S. Next-generation digital information storage in DNA. Science 337, 1628–1628 (2012).
Goldman, N. et al. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA. nature 494, 77–80 (2013).
Wang, S., Mao, X., Wang, F., Zuo, X. & Fan, C. Data storage using DNA. Adv. Mater. 36, 2307499 (2024).
Yu, M. et al. High-throughput DNA synthesis for data storage. Chem. Soc. Rev. 53, 4463–4489 (2024).
Raza, M. H., Desai, S., Aravamudhan, S. & Zadegan, R. An outlook on the current challenges and opportunities in DNA data storage. Biotechnol. Adv. 66, 108155 (2023).
De Silva, P. Y. & Ganegoda, G. U. New trends of digital data storage in DNA. Biomed Res. Int. 2016, 8072463 (2016).
DiMarco, R. L. & Heilshorn, S. C. Multifunctional materials through modular protein engineering. Adv. Mater. 24, 3923–3940 (2012).
Najafian, L. A review of bioactive peptides as functional food ingredients: mechanisms of action and their applications in active packaging and food quality improvement. Food Funct. 14, 5835–5857 (2023).
Wang, W. Lyophilization and development of solid protein pharmaceuticals. Int. J. Pharm. 203, 1–60 (2000).
Arakawa, T., Prestrelski, S. J., Kenney, W. C. & Carpenter, J. F. Factors affecting short-term and long-term stabilities of proteins. Adv. Drug Delivery Rev. 46, 307–326 (2001).
Zheng, J. et al. A mirror-image protein-based information barcoding and storage technology. Sci. Bull. 66, 1542–1549 (2021).
Hamidi, M., Azadi, A. & Rafiei, P. Hydrogel nanoparticles in drug delivery. Adv. Drug Delivery Rev. 60, 1638–1649 (2008).
Merino, S., Martín, C., Kostarelos, K., Prato, M. A. & Vázquez, E. S. Nanocomposite hydrogels: 3D polymer–nanoparticle synergies for on-demand drug delivery. ACS Nano 9, 4686–4697 (2015).
Ahmed, E. M. Hydrogel: preparation, characterization, and applications: a review. J. Adv. Res. 6, 105–121 (2015).
Vermonden, T., Censi, R. & Hennink, W. E. Hydrogels for protein delivery. Chem. Rev. 112, 2853–2888 (2012).
Bromberg, L. E. & Ron, E. S. Temperature-responsive gels and thermogelling polymer matrices for protein and peptide delivery. Adv. Drug Delivery Rev. 31, 197–221 (1998).
Koetting, M. C., Peters, J. T., Steichen, S. & Peppas, N. A. Stimulus-responsive hydrogels: Theory, modern advances, and applications. Mater. Sci. Eng. R Rep. 93, 1–49 (2015).
Yu, H. et al. Stimulus-responsive hydrogels as drug delivery systems for inflammation targeted therapy. Adv. Sci. 11, 2306152 (2024).
Fei, Z., Gupta, N., Li, M., Xiao, P. & Hu, X. Toward highly effective loading of DNA in hydrogels for high-density and long-term information storage. Sci. Adv. 9, eadg9933 (2023).
Winnik, F. M. Phase transition of aqueous poly-(N-isopropylacrylamide) solutions: a study by non-radiative energy transfer. Polymer 31, 2125–2134 (1990).
Shibayama, M. & Tanaka, T. Volume phase transition and related phenomena of polymer gels. Responsive gels: volume transitions I 1–62 (Springer Nature, 2005).
Zhang, X., Wu, D. & Chu, C. Synthesis, characterization and controlled drug release of thermosensitive IPN–PNIPAAm hydrogels. Biomaterials 25, 3793–3805 (2004).
Gupta, N., Liang, Y. N., Chew, J. W. & Hu, X. Highly robust interfacially polymerized PA layer on thermally responsive semi-IPN hydrogel: toward on-demand tuning of porosity and surface charge. ACS Appl. Mater. Interfaces 13, 60590–60601 (2021).
Alvarez-Lorenzo, C. et al. Temperature-sensitive chitosan-poly (N-isopropylacrylamide) interpenetrated networks with enhanced loading capacity and controlled release properties. J. Control. Release 102, 629–641 (2005).
Ma, F., Wang, Y. & Yang, G. The modulation of chitosan-DNA interaction by concentration and pH in solution. Polymers 11, 646 (2019).
Boonsongrit, Y., Mitrevej, A. & Mueller, B. W. Chitosan drug binding by ionic interaction. Eur. J. Pharm. Biopharm. 62, 267–274 (2006).
Ding, L., Huang, Y., Cai, X. & Wang, S. Impact of pH, ionic strength and chitosan charge density on chitosan/casein complexation and phase behavior. Carbohydr. Polym. 208, 133–141 (2019).
Bravo-Anaya, L. M., Soltero, A. R. & Rinaudo, J. F. M. DNA/chitosan electrostatic complex. Int. J. Biol. Macromol. 88, 345–353 (2016).
Huang, Z. et al. Protonated-chitosan sponge with procoagulation activity for hemostasis in coagulopathy. Bioact. Mater. 41, 174–192 (2024).
Rajabi, M., McConnell, M., Cabral, J. & Ali, M. A. Chitosan hydrogels in 3D printing for biomedical applications. Carbohydr. Polym. 260, 117768 (2021).
Wang, B. et al. Thermosensitive behavior and antibacterial activity of cotton fabric modified with a chitosan-poly (N-isopropylacrylamide) interpenetrating polymer network hydrogel. Polymers 8, 110 (2016).
Collado-González, M. et al. Chitosan as stabilizing agent for negatively charged nanoparticles. Carbohydr. Polym. 161, 63–70 (2017).
Masuoka, K. et al. The interaction of chitosan with fibroblast growth factor-2 and its protection from inactivation. Biomaterials 26, 3277–3284 (2005).
Grass, R. N., Heckel, R., Puddu, M., Paunescu, D. & Stark, W. J. Robust chemical preservation of digital information on DNA in silica with error-correcting codes. Angew. Chem. Int. Ed. 54, 2552–2555 (2015).
Wang, M. et al. Preparation and properties of chitosan-poly (N-isopropylacrylamide) semi-IPN hydrogels. J. Polym. Sci. Part A Polym. Chem. 38, 474–481 (2000).
Chen, X. et al. Preparation, characterization, and drug-release properties of pH/temperature-responsive poly (N-isopropylacrylamide)/chitosan semi-IPN hydrogel particles. J. Appl. Polym. Sci. 116, 1342–1347 (2010).
Dang, Q. et al. Controlled gelation temperature, pore diameter and degradation of a highly porous chitosan-based hydrogel. Carbohydr. Polym. 83, 171–178 (2011).
Rwei, S. P., Tuan, H. N. A., Chiang, W. Y. & Way, T. F. Synthesis and characterization of pH and thermo dual-responsive hydrogels with a semi-IPN structure based on N-isopropylacrylamide and itaconamic acid. Materials 11, 696 (2018).
Zhang, X. Z. & Chu, C. C. Fabrication and characterization of microgel-impregnated, thermosensitive PNIPAAm hydrogels. Polymer 46, 9664–9673 (2005).
Liu, Y., Lang, C., Ding, Y., Sun, S. & Sun, G. Chitosan with enhanced deprotonation for accelerated thermosensitive gelation with β-glycerophosphate. Eur. Polym. J. 196, 112229 (2023).
Zhao, S. et al. Removal of anionic dyes from aqueous solutions by adsorption of chitosan-based semi-IPN hydrogel composites. Composites Part B 43, 1570–1578 (2012).
Wu, J. Y., Liu, S. Q., Heng, P. W. S. & Yang, Y. Y. Evaluating proteins release from, and their interactions with, thermosensitive poly (N-isopropylacrylamide) hydrogels. J. Control. Release 102, 361–372 (2005).
Chen, W. D. et al. Combining data longevity with high storage capacity—layer-by-layer DNA encapsulated in magnetic nanoparticles. Adv. Funct. Mater. 29, 1901672 (2019).
Peng, Z. G., Hidajat, K. & Uddin, M. S. Adsorption of bovine serum albumin on nanosized magnetic particles. J. Colloid. Interface Sci. 271, 277–283 (2004).
Nahain, A. A., Ignjatovic, V., Monagle, P., Tsanaktsidis, J. & Ferro, V. Heparin mimetics with anticoagulant activity. Med. Res. Rev. 38, 1582–1613 (2018).
Macdougall, L. J. et al. Charged poly (N-isopropylacrylamide) nanogels for the stabilization of high isoelectric point proteins. ACS Biomater. Sci. Eng. 7, 4282–4292 (2021).
Waterman, R., Lewis, J. & Waterman, K. C. Accelerated stability modeling for peptides: a case study with bacitracin. AAPS PharmSciTech 18, 1692–1698 (2017).
Radzicka, A. & Wolfenden, R. Rates of uncatalyzed peptide bond hydrolysis in neutral solution and the transition state affinities of proteases. J. Am. Chem. Soc. 118, 6105–6109 (1996).
Milović, N. M., Dutcǎ, L. M. & Kostić, N. M. Combined use of platinum (II) complexes and palladium (II) complexes for selective cleavage of peptides and proteins. Inorg. Chem. 42, 4036–4045 (2003).
Zhang, G. & Zhu, T. F. Mirror-image trypsin digestion and sequencing of D-proteins. Nat. Chem. 16, 592–598 (2024).
Zhong, W. et al. Flow cytometry sorting for random access in DNA data storage: encapsulation for enhanced stability and sequence integrity of DNA. Anal. Chem. 40, 16099–16108 (2024).
Wang, K. et al. Unambiguous discrimination of all 20 proteinogenic amino acids and their modifications by nanopore. Nat. Methods 21, 92–101 (2024).
Zhang, Y. et al. Peptide sequencing based on host–guest interaction-assisted nanopore sensing. Nat. Methods 21, 102–109 (2024).
Zhang, M. et al. Real-time detection of 20 amino acids and discrimination of pathologically relevant peptides with functionalized nanopore. Nat. Methods 21, 609–618 (2024).
Li, G., Guo, L., Chang, X. & Yang, M. Thermo-sensitive chitosan based semi-IPN hydrogels for high loading and sustained release of anionic drugs. Int. J. Biol. Macromol. 50, 899–904 (2012).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant 22477075, 22077078) and the National Key R&D Program of China (2021YFC2100201).
Author information
Authors and Affiliations
Contributions
B. Luo: Investigation, Methodology, Software, Data curation, Writing - original draft. S. Gao and M. Wu: Investigation, Methodology. X. Dong: Conceptualization, Supervision, Resources, Project administration. X. Deng and H. Hu: Conceptualization, Supervision, Resources, Writing-Reviewing and Editing, Project administration, Funding acquisition.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Materials thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Youn Soo Kim and Jet-Sing Lee. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Luo, B., Gao, S., Wu, M. et al. High-capacity information storage using peptide-encapsulated hydrogels for long-term data preservation. Commun Mater 6, 183 (2025). https://doi.org/10.1038/s43246-025-00915-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s43246-025-00915-y