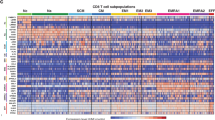
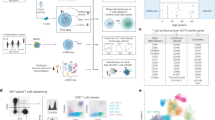
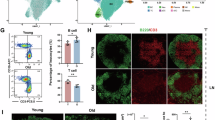
Abstract
Aging is characterized by the progressive deterioration of tissue structure and function, leading to increased vulnerability to diseases. Senescent cells (SCs) accumulate with age, but how the immune system regulates their burden is unclear. Here we show that CD4 T cells differentiate into Eomesodermin (Eomes)+CCL5+ T lymphocytes (CD4-Eomes) in a SC-rich environment and that a reduction in the SC load, achieved using senolytic drugs, was sufficient to halt this differentiation. We further demonstrate that eliminating CD4-Eomes cells at advanced age by selectively deleting the Eomes transcription factor in CD4 T cells results in increased accumulation of SCs, profound physical deterioration and a decreased lifespan. In liver cirrhosis, a model of localized chronic inflammation, CD4-Eomes cell elimination increased fibrosis, SC load and worsened the disease. Collectively, our findings demonstrate the fundamental role of CD4-Eomes cells in modulating tissue senescence, with implications for age-related diseases and longevity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data supporting the findings of this study are included in the figures and supplementary information files of the manuscript. All data supporting the findings of the study are available from the corresponding author upon reasonable request.
References
López-Otín, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. Hallmarks of aging: an expanding universe. Cell 186, 243–278 (2023).
Pathak, R. U., Soujanya, M. & Mishra, R. K. Deterioration of nuclear morphology and architecture: a hallmark of senescence and aging. Ageing Res. Rev. 67, 101264 (2021).
Di Micco, R., Krizhanovsky, V., Baker, D. & d’Adda di Fagagna, F. Cellular senescence in ageing: from mechanisms to therapeutic opportunities. Nat. Rev. Mol. Cell Biol. 22, 75–95 (2021).
Vilas, J. M. et al. Adult Sox2+ stem cell exhaustion in mice results in cellular senescence and premature aging. Aging Cell 17, e12834 (2018).
Yousefzadeh, M. J. et al. An aged immune system drives senescence and ageing of solid organs. Nature 594, 100–105 (2021).
Mogilenko, D. A. et al. Comprehensive profiling of an aging immune system reveals clonal GZMK+ CD8+ T cells as conserved hallmark of inflammaging. Immunity 54, 99–115 (2021).
Abdellatif, M., Rainer, P. P., Sedej, S. & Kroemer, G. Hallmarks of cardiovascular ageing. Nat. Rev. Cardiol. 20, 754–777 (2023).
Zhang, P. et al. Senolytic therapy alleviates Aβ-associated oligodendrocyte progenitor cell senescence and cognitive deficits in an Alzheimer’s disease model. Nat. Neurosci. 22, 719–728 (2019).
Saez-Atienzar, S. & Masliah, E. Cellular senescence and Alzheimer disease: the egg and the chicken scenario. Nat. Rev. Neurosci. 21, 433–444 (2020).
López-Otín, C., Pietrocola, F., Roiz-Valle, D., Galluzzi, L. & Kroemer, G. Meta-hallmarks of aging and cancer. Cell Metab. 35, 12–35 (2023).
Kolodkin-Gal, D. et al. Senolytic elimination of Cox2-expressing senescent cells inhibits the growth of premalignant pancreatic lesions. Gut 71, 345–355 (2022).
Chaib, S., Tchkonia, T. & Kirkland, J. L. Cellular senescence and senolytics: the path to the clinic. Nat. Med. 28, 1556–1568 (2022).
Childs, B. G., Durik, M., Baker, D. J. & van Deursen, J. M. Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nat. Med. 21, 1424–1435 (2015).
Covarrubias, A. J. et al. Senescent cells promote tissue NAD+ decline during ageing via the activation of CD38+ macrophages. Nat. Metab. 2, 1265–1283 (2020).
Yosef, R. et al. Directed elimination of senescent cells by inhibition of BCL-W and BCL-XL. Nat. Commun. 7, 11190 (2016).
Campisi, J. & d’Adda di Fagagna, F. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 8, 729–740 (2007).
Rodier, F. et al. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat. Cell Biol. 11, 973–979 (2009).
Ovadya, Y. et al. Impaired immune surveillance accelerates accumulation of senescent cells and aging. Nat. Commun. 9, 5435 (2018).
Ovadya, Y. & Krizhanovsky, V. Strategies targeting cellular senescence. J. Clin. Invest. 128, 1247–1254 (2018).
Kang, T.-W. et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature 479, 547–551 (2011).
Krizhanovsky, V. et al. Senescence of activated stellate cells limits liver fibrosis. Cell 134, 657–667 (2008).
Pereira, B. I. et al. Senescent cells evade immune clearance via HLA-E-mediated NK and CD8+ T cell inhibition. Nat. Commun. 10, 2387 (2019).
Mittelbrunn, M. & Kroemer, G. Hallmarks of T cell aging. Nat. Immunol. 22, 687–698 (2021).
Carrasco, E. et al. The role of T cells in age-related diseases. Nat. Rev. Immunol. 22, 97–111 (2022).
Elyahu, Y. et al. Aging promotes reorganization of the CD4 T cell landscape toward extreme regulatory and effector phenotypes. Sci. Adv. 5, eaaw8330 (2019).
Patil, V. S. et al. Precursors of human CD4+ cytotoxic T lymphocytes identified by single-cell transcriptome analysis. Sci. Immunol. 3, eaan8664 (2018).
Soto-Heredero, G., Gómez de Las Herass, M. M., Escrig-Larena, J. I. & Mittelbrunn, M. Extremely differentiated T cell subsets contribute to tissue deterioration during aging. Annu. Rev. Immunol. 41, 181–205 (2023).
Zuroff, L. et al. Immune aging in multiple sclerosis is characterized by abnormal CD4 T cell activation and increased frequencies of cytotoxic CD4 T cells with advancing age. EBioMedicine 82, 104179 (2022).
Kaneko, N. et al. Temporal changes in T cell subsets and expansion of cytotoxic CD4+ T cells in the lungs in severe COVID-19. Clin. Immunol. 237, 108991 (2022).
Joulia, E. et al. Eomes-dependent mitochondrial regulation promotes survival of pathogenic CD4+ T cells during inflammation. J. Exp. Med. 221, e20230449 (2024).
Hashimoto, K. et al. Single-cell transcriptomics reveals expansion of cytotoxic CD4 T cells in supercentenarians. Proc. Natl Acad. Sci. USA 116, 24242–24251 (2019).
Hasegawa, T. et al. Cytotoxic CD4+ T cells eliminate senescent cells by targeting cytomegalovirus antigen. Cell 186, 1417–1431 (2023).
Eshima, K. et al. Ectopic expression of a T-box transcription factor, eomesodermin, renders CD4+ Th cells cytotoxic by activating both perforin- and FasL-pathways. Immunol. Lett. 144, 7–15 (2012).
Curran, M. A. et al. Systemic 4-1BB activation induces a novel T cell phenotype driven by high expression of Eomesodermin. J. Exp. Med. 210, 743–755 (2013).
Desdín-Micó, G. et al. T cells with dysfunctional mitochondria induce multimorbidity and premature senescence. Science 368, 1371–1376 (2020).
Dimri, G. P. et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl Acad. Sci. USA 92, 9363–9367 (1995).
Bhattacharya, M. & Ramachandran, P. Immunology of human fibrosis. Nat. Immunol. 24, 1423–1433 (2023).
De Cecco, M. et al. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 566, 73–78 (2019).
Del Rey, M. J. et al. Senescent synovial fibroblasts accumulate prematurely in rheumatoid arthritis tissues and display an enhanced inflammatory phenotype. Immun. Ageing 16, 29 (2019).
Schafer, M. J. et al. Cellular senescence mediates fibrotic pulmonary disease. Nat. Commun. 8, 14532 (2017).
Amor, C. et al. Senolytic CAR T cells reverse senescence-associated pathologies. Nature 583, 127–132 (2020).
Ogrodnik, M. et al. Cellular senescence drives age-dependent hepatic steatosis. Nat. Commun. 8, 15691 (2017).
Nallagangula, K. S., Nagaraj, S. K., Venkataswamy, L. & Chandrappa, M. Liver fibrosis: a compilation on the biomarkers status and their significance during disease progression. Future Sci. OA 4, FSO250 (2018).
McPherson, S., Stewart, S. F., Henderson, E., Burt, A. D. & Day, C. P. Simple non-invasive fibrosis scoring systems can reliably exclude advanced fibrosis in patients with non-alcoholic fatty liver disease. Gut 59, 1265–1269 (2010).
Wai, C.-T. et al. A simple noninvasive index can predict both significant fibrosis and cirrhosis in patients with chronic hepatitis C. Hepatology 38, 518–526 (2003).
Ogrodnik, M. & Gladyshev, V. N. The meaning of adaptation in aging: insights from cellular senescence, epigenetic clocks and stem cell alterations. Nat. Aging 3, 766–775 (2023).
Lau, V., Ramer, L. & Tremblay, M.-E. An aging, pathology burden, and glial senescence build-up hypothesis for late onset Alzheimer’s disease. Nat. Commun. 14, 1670 (2023).
Giannoula, Y., Kroemer, G. & Pietrocola, F. Cellular senescence and the host immune system in aging and age-related disorders. Biomed. J. 46, 100581 (2023).
Bitencourt, T. C., Vargas, J. E., Silva, A. O., Fraga, L. R. & Filippi-Chiela, E. Subcellular structure, heterogeneity, and plasticity of senescent cells. Aging Cell 23, e14154 (2024).
Oh, D. Y. & Fong, L. Cytotoxic CD4+ T cells in cancer: expanding the immune effector toolbox. Immunity 54, 2701–2711 (2021).
Meckiff, B. J. et al. Imbalance of regulatory and cytotoxic SARS-CoV-2-reactive CD4+ T cells in COVID-19. Cell 183, 1340–1353 (2020).
Cenerenti, M., Saillard, M., Romero, P. & Jandus, C. The era of cytotoxic CD4 T cells. Front. Immunol. 13, 867189 (2022).
Raveney, B. J. E. et al. Involvement of cytotoxic Eomes-expressing CD4+ T cells in secondary progressive multiple sclerosis. Proc. Natl Acad. Sci. USA 118, e2021818118 (2021).
Macy, A. M., Herrmann, L. M., Adams, A. C. & Hastings, K. T. Major histocompatibility complex class II in the tumor microenvironment: functions of nonprofessional antigen-presenting cells. Curr. Opin. Immunol. 83, 102330 (2023).
Zindl, C. L. et al. Distal colonocytes targeted by C. rodentium recruit T-cell help for barrier defence. Nature 629, 669–678 (2024).
Brabec, T. et al. Segmented filamentous bacteria-induced epithelial MHCII regulates cognate CD4+ IELs and epithelial turnover. J. Exp. Med. 221, e20230194 (2024).
Majewska, J. et al. p16-dependent increase of PD-L1 stability regulates immunosurveillance of senescent cells. Nat. Cell Biol. 26, 1336–1345 (2024).
Oh, D. Y. et al. Intratumoral CD4+ T cells mediate anti-tumor cytotoxicity in human bladder cancer. Cell 181, 1612–1625 (2020).
Cachot, A. et al. Tumor-specific cytolytic CD4 T cells mediate immunity against human cancer. Sci. Adv. 7, eabe3348 (2021).
Juno, J. A. et al. Cytotoxic CD4 T cells—friend or foe during viral infection? Front. Immunol. 8, 19 (2017).
Helman, A. et al. p16Ink4a-induced senescence of pancreatic beta cells enhances insulin secretion. Nat. Med. 22, 412–420 (2016).
Acknowledgements
The study was supported by the Ministry of Science and Technology (grant no. 3-16148) and the Litwin and Gural Foundations. We thank A. Tarasiuk and A. Rudich for their guidance and support with the metabolic cages platform. We thank I. Ben-Porath (Hebrew University, Israel) for providing tissue sections from p16-deficient mice, and we thank M. Schiller (Tel-Aviv University, Israel) for her valuable comments on the manuscript.
Author information
Authors and Affiliations
Contributions
Y.E. and A.M. conceived the project, designed the experiments and wrote the manuscript. I.F., N.P., A.Z., A.S., O.B., R.A.-M., E.E., A.N. and K.R. performed the experimental work and analyzed the data. L.R. and V.K. provided technical and scientific support.
Corresponding author
Ethics declarations
Competing interests
A.M. and Y.E. (‘Immune system restoration by cell therapy’) have a pending US patent application (no. 18/043576), a pending EU patent application (no. 21863834.4) and a pending Israel patent application (no. 301045). A related specific aspect of the manuscript covered in this patent application is the emphasis on CD4-Eomes T cells as a potential biomarker of cellular senescence and treatment of aging and age-related diseases involving SC accumulation and immune system restoration. The other authors declare no competing interests.
Peer review
Peer review information
Nature Aging thanks Shadmehr Demehri, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Adoptively transferred young-origin CD4 T cell differentiation in an aged environment.
a. Representative flow cytometry plots showing the gating strategy used to define CD4 T cell subsets. b. Representative flow cytometry plots showing phenotypic characteristics of the young CD45.1 cells used for adoptive transfer experiments. c. Quantitative analysis of CD4+ cells among the transferred cells in young (n = 6) or old (n = 4) WT mice (CD45.2). d. Histogram plots showing fluorescence of CFSE-labeled CD45.1+CD4+ cells relative to unlabeled cells. e. Graph showing the analysis of transferred cells (CD45.1+); the percentage of CD4+CD44+ T cells out of CFSE− or CFSE+ cells in old mice (CD45.2; n = 6). f. Representative confocal images showing γH2A.X immunostaining in the Vehicle (Left) and Senolytic (Right) groups. Scale bars are indicated in the figures. g. Representative immunofluorescence images showing liver sections of Vehicle mice immunolabeled with a secondary antibody as a negative control (Lower) or stained with DAPI (Upper). Scale bars are indicated in the figures. h-i. The percentages of Treg (FOXP3+, Left), naïve (CD3+CD4+CD62L+CD44−, Middle), effector (CD3+CD4+CD62L−CD44+, Right) (h) and PD1+ (CD3+CD4+CD62L−CD44+PD1+) (i) CD4+ T cells originating from CD45.1 young mice and transferred into CD45.2 old control (n = 11) and senolytic drug-treated (n = 10) mice. Each dot represents the percentages in a single mouse. Bars represent mean ± SEM from two (c and e) or three (h-i) independent experiments. Data were analyzed using two-tailed unpaired (c, h-i) and paired (e) Student’s t-tests.
Extended Data Fig. 2 Comparison of endogenous (CD45.2) and transferred (CD45.1) subsets in senolytic-treated old mice.
a. Graph shows the percentages of EOMES⁺CCL5⁺ cells originating from CD45.1+ cells in control (n = 11) and senolytic drug-treated (n = 10) mice. b-e. The ratio between young-transferred (CD45.1) and old-endogenous (CD45.2) Treg (CD4+FOXP3+; b), naïve (CD3+CD4+CD62L+CD44−; c), effector (CD3+CD4+CD62L−CD44+; d), and CD4+ PD1+ (e) cells in old control (Control; n = 11) and senolytic drug-treated mice (Senolytic; n = 10). Each dot represents the ratio in a single mouse. The right panels show the percentage of old-endogenous (CD45.2) and the young-transferred (CD45.1) cells of each subset in the old-control (Left) and old-senolytic (Right) mice. Bars represent mean ± SEM from three independent experiments. Data were analyzed using two-tailed unpaired (Left panels) or paired (Right panels) Student’s t-tests.
Extended Data Fig. 3 Senescent cells induce CD4-Eomes cell differentiation in vitro.
a. Experimental setup: primary fibroblasts derived from mouse lungs were treated with etoposide to induce senescence (Methods). Both control untreated and senescent fibroblasts were then co-cultured with polyclonally activated young CD4 T cells (purified from 2-3 months old mice) for 3 or 7 days when CD4 T cells were collected for flow cytometry analysis. b. Representative images (Left panels) of senescence-associated β-Galactosidase (SA-β-Gal) staining in control (Upper) and etoposide-induced senescent fibroblasts (Lower). Scale bar represents 50 μm. Right: a graph showing the percentages of SA-β-Gal+ cells out of total fibroblasts under 40x magnification in control (n = 5) and etoposide-induced senescent fibroblasts (n = 5) (Etoposide). Each dot represents one culture well. c. Representative confocal images showing P16 and P21 immunostaining in control (Upper) and senescent (Lower) fibroblasts. Below to the left is a validation of the P16 antibody specificity on liver tissue sections obtained from P16 deficient mice (3 months of age) and immunostained with the P16 antibody and DAPI. Scale bars are indicated in the figures. Data from ≥4 wells d-e. Left: a representative flow cytometry histogram plot illustrating the expression of EOMES in CD4 T cells harvested from non-activated control (grey), activated with control fibroblasts (light green), or activated with senescent fibroblasts (blue), after 3 days of co-culture. Right: Fold change (FC) of median fluorescence intensity (MFI) analysis of EOMES in activated CD4 T cells co-cultured with either control (n = 9) or senescent (Sc) fibroblasts (n = 9) after 3 (d) or 7 (e) days of co-culture. Each dot represents one well. f-g. FC of GzmB MFI (Left) and percentage of GzmB+ cells (Right) out of activated CD4 T cells co-cultured with control (n = 9) or senescent fibroblasts (n = 9), after 3 (f) or 7 (g) days of co-culture. h-i. FC of PD1 (Left) and CD44 (Right) MFI in activated CD4 T cells co-cultured with control (PD1:n = 9, CD44:n = 6) or senescent fibroblasts (PD1:n = 9; CD44:n = 6), after 3 (h) or 7 (i) days of co-culture. Each value was normalized to the mean MFI values of the control group. j. Experimental setup: polyclonally activated young CD4 T cells were cultivated with supernatant from control or senescent fibroblasts for 3 days and then collected for flow cytometry analysis. k. FC of EOMES MFI in non-activated (NA) or activated CD4 T cells cultivated with supernatants from control (n = 6) or senescent cells (n = 6). l-m. Representative confocal images showing DAPI staining together with MHCII (l) or ICAM-1(m) immunostaining in control (Upper) and Sc (Lower) fibroblasts. Data from ≥4 well. Scale bars are indicated in the figures. Bars represent mean ± SEM from two independent experiments. Data were analyzed using a two-tailed Student’s t-test, unpaired, with exact P-values presented in the graphs. Created with BioRender.com.
Extended Data Fig. 4 Validation of the CD4-CreERT2+/−Eomesfl/fl mouse model.
Splenocytes from either CD4-CreERT2+/−Eomesfl/fl mice (Eomes-KO; n = 5; age 3 months-old) or CD4-CreERT2−/−Eomesfl/fl mice (Control; n = 5) were isolated and analyzed for EOMES+ cells out of spleen-derived CD4 T cells with flow cytometry. In addition, spleen-derived T cells were activated using anti-CD3/anti-CD28 coated beads for 24 h and analyzed for cell survival and cytokine production with flow cytometry. a. Left: percentage of CD4 T cells (CD3+CD8−CD4+); Right: CD4 MFI. b. Left: percentage of live cells after activation; Right: percentage of CD4 T cells out of the live cells. c. Left: percentage of EOMES+ cells out of CD8 T cells; Right: quantitative analysis of EOMES MFI in CD8 T cells. d. Left: percentage of EOMES+ cells out of CD4 T cells; Middle: quantitative analysis of EOMES MFI in CD4 T cells; Right: a representative flow cytometry histogram plot illustrating the expression of EOMES in non-activated CD4 T cells (grey), activated Eomes-KO CD4 T cells (blue), or activated control CD4 T cells (orange). e. ELISA results of IFNγ (Left) and IL-2 (Right) in the supernatants of CD4 T cells derived from Control (n = 5) or Eomes-KO (n = 5) mice and activated for 24 h. Bars indicate mean ± SEM. Data were analyzed using a two-tailed Student’s t-test, unpaired, with exact P-values presented in the graphs.
Extended Data Fig. 5 CD4/Eomes/CCL5 T cell frequencies in control and Eomes-KO mice (a-b); the metabolic performance of control and Eomes-KO mice before TMX administration (c-i).
a. Graphs showing CD4+CCL5+ in the blood before and after TMX treatment in the control (Left, n = 6) and in the Eomes-KO (Right, n = 7) groups. b. Validation of TMX effect on frequencies of CD4 and CD4-EOMES T cells: graphs show the percentages of CD4 T cells (Left) and CD4-EOMES cells (Right) after vehicle (n = 5) or TMX treatment (n = 4) in young (4-5 months old) WT mice. Regimen consisted of intraperitoneal injections of 100 µl of TMX for three days followed by an alternating dietary regimen, where TMX chow and regular chow were alternated every two weeks, for a total duration of six weeks. c. Experimental Setup: 20-month-old Control (CreERT2−/−Eomesfl/fl) and Eomes-KO (CreERT2+/−Eomesfl/fl) mice were evaluated in metabolic cages for a week before the TMX regiment was administered. d. Representative temperature recording during the experiment. e-i. Representative comparison between Control (n = 3) and Eomes-KO (n = 3) groups for various parameters recorded over 312 h at 30-minute intervals. These parameters include fine activity measured by x-axis beam breaks (e), energy expenditure (kcal/hour; f), food consumption (grams; g) water consumption (grams; h) and distance traveled (meters; i). Bars indicate mean ± SEM. Data were analyzed using a two-tailed Student’s t-test, paired (a) or unpaired (b), from one experiment, with exact P-values presented in the graphs. Created with BioRender.com.
Extended Data Fig. 6 Metabolic cage performance before and after tamoxifen administration in control and Eomes-KO mice.
a. The death rate in the Control group (30%) versus the Eomes-KO group (70%) during the time of metabolic experiments (from the first to the second metabolic recording, a 45-day period). b-e. Graphs showing weight (grams; b), activity (meters; c), food intake (grams; d), and water intake (grams; e) before and after TMX treatment in the Control (Left; n = 12) and in the Eomes-KO (Right; n = 14) groups. Data were analyzed using two-tailed paired Student’s t-tests. Data from two independent experiments.
Extended Data Fig. 7 Analysis of CD4 T-cell subsets in the liver and spleen of control and Eomes-KO mice.
a. The percentages of CD4-EOMES (Left) and CD4+GzmB+ cells (Right) in spleens of Control (n = 6) and Eomes-KO (n = 7) mice after the TMX regimen was administered. The regimen included 3 days of IP TMX injection (100 µl) followed by an alternating dietary regimen of two weeks on TMX chow and two weeks on regular chow, lasting for 6 weeks. b-e. The percentages of CD45+ (b; Left), CD45+CD3+ (b; Right), CD3+CD8+ (c; Left), CD3+CD4+ (c; Right), naive (d; Left), effector (d; Right), Treg (e; Left), and CD44+PD1+ (e; Right) cells in the livers of Control (n = 6) and Eomes-KO mice (n = 7). f-h. Graphs showing the percentages of CD3+ (f; Left), CD3+CD4+ (f; Right), Treg (g; Left), naive (g; Right), effector (h; Left), and CD44+PD1+ (h; Right) cells in the spleens of Control (n = 6) or Eomes-KO mice (n = 7). Subsets were defined as indicated in Supplementary Figure 1. Data were analyzed using a two-tailed, unpaired Student’s t-test. The exact P-values are presented in the graphs. i. A representative flow cytometry histogram plot illustrating the expression of β-Galactosidase (β-Gal) staining in liver cells (light purple), unstained liver cells (grey) and Fluorescence Minus One Controls (FMO) liver cells (light blue). j. A representative flow cytometry histogram plot illustrating the expression of p16ink4a+ (Upper) and p21 (Lower) out of non-immune CD45− liver cells. k-l. The percentages of CD45+p16+ (k; Left), fold change of p16 MFI (k; Right), percentages of CD45+p21+ (l; Left), fold change of p21MFI (l; Right) in the livers of Control (n = 6) or Eomes-KO mice (n = 7). m. Tracking of mice weight (Y-axis) over 40 weeks (X-axis) in the Control and Eomes-KO groups (n ≥ 4 mice per time point). * = P < 0.05. n-r. Survival experiment: 18 month-old control and Eomes-KO mice were subjected to a TMX regimen that included 3 days of IP TMX injection (100 µl) followed by an alternating dietary regimen of two weeks on TMX chow and two weeks on regular chow, lasting for 30 weeks. n. Representative spleen size of Control and Eomes-KO mice. o. Comparison of spleen weight (mg) in Control (n = 4) and Eomes-KO (n = 6) mice. p-q. The percentages of CD3 cells (Left; p), CD4 cells (Right; p) and CD4-EOMES cells (q) in spleens of Control (n = 4) and Eomes-KO mice (n = 6). r. Representative flow cytometry plots showing CD4+EOMES+CCL5+ cells in the spleen of Control (Upper) and Eomes-KO mice (Lower) after TMX treatment. Bars indicate mean ± SEM from one (a-h, k-l) or two (m-r) independent experiments. Data were analyzed using two-tailed Student’s t-tests. The exact P-values are presented in the figures.
Extended Data Fig. 8 Systemic immune response to CCl4-induced liver inflammation in control and Eomes-KO mice.
a-b. Weight (Y-axis) of TMX-Control (n = 12), CCl4-Control (n = 12), and CCl4-Eomes-KO (n = 15) mice measured over 6 weeks (X-axis) (a), and the percentages of CD3+CD4+ cells in their livers (b). c-d. The percentages of CD3+CD4+, CD3+CD4+FOXP3+ Treg, CD44+CD62L− effector, and CD44+CD62L−PD1+ cells (left to right) in the blood (c) or in the spleen (d) of TMX-control (n = 12), CCL4 -control (n = 12), CCL4-Eomes-KO (n = 15) mice. e. Representative immunofluorescence images showing liver sections of TMX-control immunolabeled with p21 (Lower) or stained with DAPI (Upper). f. Representative immunofluorescence images showing liver sections of CCl4-Eomes-KO mice immunolabeled with a secondary antibody as a negative control (Lower) or stained with DAPI (Upper), Scale bars are indicated in the figures. Bars indicate mean ± SEM from two independent experiments (a-d). Data were analyzed using one-way ANOVA with Tukey correction for multiple comparisons. Exact P-values are presented in the figures. “ns” denotes non-significant across all time-points in (a).
Extended Data Fig. 9 Senolytic treatment mitigates liver fibrosis and senescent cell load alongside reduced accumulation of Eomes-CD4 T cells in a mouse model of liver cirrhosis.
a. Experimental setup: Young (4-5 months old) WT mice were treated with CCl4 (CCl4-Control), CCl4 combined with senolytic (CCl4-Senolytic), or vehicle control (Control) for 45 days. Spleens, blood, and livers were collected for further analysis. b. Representative liver samples from CCl4-Control (Upper) and CCl4-Senolytic (Lower) mice showing reduced scarring (marked by yellow arrows). Estimated scale bar represents 5 mm. c. Left: representative liver tissue images from CCl4-Control (Upper) and CCl4-Senolytic (Lower) mice, stained with Sirius Red. The scale bar represents 50 μm. Right: Pie plots showing the percentages of liver fibrosis severity scores (A-D) for CCl4-Control (n = 7, Upper), and CCl4-Senolytic (n = 7; Lower) mice (Methods). d. Quantitative analysis of Sirius Red staining area calculated as the percentage of tissue area strained with Sirius Red out of all tissue area (ROI) for Control (n = 4), CCl4-Control (n = 7), and CCl4-Senolytic (n = 7) mice. Each dot represents the average of at least 2 sections of liver tissue from one mouse. e. Representative confocal images of γH2A.X (Upper) and P21 (Lower) immunostaining and quantification in the CCl4-Control (n = 5) and CCl4-Senolytic groups (n = 5). Scale bars are indicated in the figures. Left panels- graph shows quantitative analysis of γH2A.X and P21, calculated as γH2A.X+DAPI+ or P21+DAPI+ cells out of DAPI+ cells. Each dot represents the average of at least 2 sections per individual mouse. f. The percentages of CD4-EOMES cells in the livers (Left), spleens (Middle), and blood samples (Right) of Control (n = 4), CCl4-Control (n = 7), and CCl4-Senolytic (n = 7) mice. g. Quantitative analysis of GzmB MFI in CD4 T cells from the livers of Control (n = 4), CCl4-Control (n = 7), and CCl4-Senolytic (n = 7) mice. h. Graphs showing the percentages (left to right) of CD4+, Treg, effector, and CD44+PD1+ cells in the spleens of Control (n = 4), CCl4-Control (n = 7), and CCl4-Senolytic mice (n = 7). Bars indicate mean ± SEM from two (a-d; f-h) or one (e) independent experiments. Data were analyzed using one-way ANOVAs with Tukey correction for multiple comparisons. Each dot represents one mouse; the p-value is indicated in the figures. Created with BioRender.com.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Elyahu, Y., Feygin, I., Eremenko, E. et al. CD4 T cells acquire Eomesodermin to modulate cellular senescence and aging. Nat Aging 5, 1970–1982 (2025). https://doi.org/10.1038/s43587-025-00953-8
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s43587-025-00953-8