Abstract
Background
Liquid biopsy approaches, especially the detection of circulating tumor DNA (ctDNA), are emerging as sensitive and reliable surrogates for tumor tissue-based routine diagnostic testing. Here, we retrospectively analyzed serially collected plasma samples of non-small cell lung cancer (NSCLC) patients obtained at first diagnosis to evaluate the added value of ctDNA analysis for detecting therapeutically relevant variants and determining the consequent clinical implications.
Methods
One hundred eighty plasma samples from consecutively recruited NSCLC patients were included. Circulating cell-free DNA (ccfDNA) was extracted and analyzed with the UltraSEEK Lung Panel v2 on the MassARRAY System. Tumor tissue next-generation sequencing (NGS) data, performed as routine molecular testing in the clinical setting, were retrieved from the national pathology registry for 132 patients.
Results
Here we show that in 82% of the patients, mutations are concordantly detected in tumor tissue and plasma. More mutations are reported with tumor tissue-based NGS in nineteen patients, while in four patients additional mutations are detected in plasma. Tissue-based molecular tumor profiling identifies 60 patients eligible for targeted treatment including fifteen (8%) harboring fusions currently not covered by UltraSEEK. Based on ctDNA analysis, 41 patients (23%) are identified as eligible for BRAFV600-, EGFR-, or KRASG12C-targeted therapies. In the absence of tumor tissue NGS data (n = 48), five therapeutically relevant mutations are detected.
Conclusions
Molecular tumor profiling of ctDNA identifies therapeutically relevant mutations at a comparable rate to tumor tissue-based NGS and might therefore serve as an alternative or complementary test for the detection of actionable variants in plasma.
Plain language summary
Being able to identify cancer mutations to direct treatment strategies has become very important in improving outcomes of patients with non-small cell lung cancer. With the detection of cancer DNA in the blood, referred to as liquid biopsy, an alternative to invasive tissue-based diagnostics now exists. We tested how sensitive the UltraSEEK Lung Panel (a method which evaluates a panel of genes) was in the detection of mutations in blood affecting treatment decisions for patients with non-small cell lung cancer. The same mutations were detected using tissue and this blood-based method in 82% of the patients. Moreover, in a similar number of patients, the most favorable treatment advice was based on tissue and blood information. This study showed that blood testing is suitable for the detection of mutations and has the potential to improve diagnostics for lung cancer patients.
Similar content being viewed by others
Introduction
The mutational spectrum of non-small cell lung cancer (NSCLC) is remarkably heterogeneous, wherein many single nucleotide variants (SNVs), insertions or deletions (indels), fusions, genomic rearrangements, and copy number alterations (CNAs) can occur within the tumor genome1. Current international guidelines recommend testing for molecular aberrations for patients with metastatic lung adenocarcinoma. In this tumor type, pathogenic mutations can be detected in > 60% of the cases2,3. To date, targeted therapies are available for mutations in BRAF (affecting valine at codon 600), EGFR, ERBB2, KRAS [specifically p.(G12C)] and MET (resulting in exon 14 skipping), as well as for fusions of ALK, NRG1, NTRK1, NTRK2, NTRK3, RET, and ROS14,5. In the Netherlands, molecular profiling of common aberrations in these twelve predictive genes is recommended using both RNA- and DNA-based next-generations sequencing (NGS) panels6. BRAF, EGFR and KRAS variants have a combined prevalence of ~ 50% in advanced-stage lung adenocarcinoma2. Although therapeutically targetable mutations in BRAF, EGFR and KRAS are detected in 30–35% of all lung adenocarcinoma patients, these variants represent > 80% of all the actionable mutations reported in clinical practice2,4. Genomic alterations, not limited to targetable alterations, occur significantly less frequent in other NSCLC entities, including lung squamous cell carcinoma (<25%) and not otherwise specified (NOS) lung carcinoma (~ 40%)2,7. In order to obtain high sensitivity to detect (actionable) variants in tumor DNA, NGS analyses require a sufficient amount and percentage of neoplastic cells. In addition, extracted DNA and RNA need to be of adequate quantity and quality. Tumor tissue specimens in metastatic NSCLC are frequently collected through endobronchial ultrasound-guided transbronchial needle aspiration (EBUS-TBNA) or transesophageal ultrasound-guided fine needle aspiration (EUS-FNA), but these methods generally yield little tumor tissue8,9. In 20–30% of lung cancer patients, tumor sampling is not possible or NGS results fail due to low tumor DNA quantity8,10,11,12. Therefore, there is an urgent need for alternative methods to determine the predictive biomarker status for treatment decision making.
Molecular profiling of circulating cell-free DNA (ccfDNA) has proven to be a reliable alternative method to detect tumor-derived mutations in lung cancer patients10,13. Circulating tumor DNA (ctDNA) represents only a small fraction of the ccfDNA (often less than 1%) and gradually lower in earlier stages of disease14. Therefore, ctDNA detection assays with high sensitivity and specificity have been developed. Clinical applications of ctDNA testing include baseline predictive biomarker identification and resistance mutation detection at disease progression, particularly when tumor tissue is inadequate15. Several ctDNA-based liquid biopsy approaches are designed as single target assays (e.g., droplet digital PCR (ddPCR)), single or multiple gene panels (e.g., cobas®, Idylla™, UltraSEEK) or elaborate NGS panels (e.g., Oncomine, AVENIO™, FoundationOne® Liquid CDx, Guardant360® CDx)16,17. Although high sensitivities and specificities can be achieved with these approaches, the accuracy of molecular tumor profiling generally increases as more targets are analyzed simultaneously18,19. The choice of assay, however, depends not only on the coverage of targets but also on costs, eligibility for reimbursement, turnaround time, availability of patient-derived material and the availability of technical expertise to operate a testing platform20.
Mid-sized targeted liquid biopsy panels covering most actionable mutations observed in lung adenocarcinoma are commercially available. These panels have considerably lower costs and processing times compared to both tumor tissue- and plasma-based NGS approaches without compromising on analytical sensitivity20,21. Therefore, a highly sensitive, cost-effective liquid biopsy test with a short turnaround time might provide valuable information on the (therapeutically targetable) mutation status of the tumor and assist in treatment decision making11. For the implementation of liquid biopsy approaches in routine diagnostics, pre-treatment testing of plasma samples simultaneously collected as tumor tissue specimens is necessary to demonstrate its clinical value.
The aim of this study was to retrospectively evaluate the added value of ctDNA analysis for the detection of therapeutically relevant variants in an unselected series of lung cancer patients. For this purpose, pre-treatment plasma samples of advanced-stage NSCLC patients were serially collected without upfront knowledge of the availability of lung cancer tissue biopsy specimens and the results of its molecular profile. For each patient, treatment decisions based on (actionable) mutations identified in tumor tissue DNA or ctDNA were determined in retrospect in accordance with current international guidelines5. The ctDNA was analyzed using the UltraSEEK Lung Panel on the MassARRAY System, which has recently demonstrated high concordance with tumor tissue NGS in selected populations, particularly for therapeutically targetable mutations22,23,24. To resemble a real-world clinical setting in this study, a continuous diagnostic workflow from ccfDNA extraction to an UltraSEEK result was applied and time-to-process was determined.
Methods
Patient inclusion and sample processing
Between June 2018 and December 2021, pre-treatment plasma samples of 180 consecutive patients treated for NSCLC at the University Medical Center Groningen (UMCG) were obtained from the OncoLifeS Biobank. Inclusion criteria consisted of (1) histologically confirmed NSCLC, (2) no previous cancer-related treatment, (3) sufficient plasma for ccfDNA extraction according to standard operating procedures, and 4) patients were not recruited for a specific study to prevent selection bias. The use of patient-derived plasma samples for research purposes was approved by the Medical Ethical Committee (METc) of the UMCG (2010/109), as well as for this study (Reg nr. 202200300); molecular analyzes were performed in a NEN-EN-ISO 15189-accredited laboratory. All patients provided written informed consent.
Pre-treatment plasma samples were collected in Cell-Free DNA BCTs® (Streck, Omaha, NE, USA). Blood samples were processed within 48 hours. Blood collection tubes (BCTs) were centrifuged at 1600 × g for 10 minutes; the supernatant was subsequently centrifuged for 10 minutes at 16,000 × g22,25. Cell-free plasma was stored in 1 mL fractions at −80 °C until ccfDNA extraction. ccfDNA was extracted from 2 mL of cell-free plasma and eluted in 47 μL AVE elution buffer with QiaAMP Circulating Nucleic Acid Kit (Qiagen) according to the manufacturer’s recommendations26. Extracted ccfDNA was quantified using both the Qubit dsDNA HS Assay (Thermo Fisher Scientific, Waltham, MA, USA) and the LiquidIQ® Panel (Agena Bioscience) following the manufacturer’s instructions22,27, which correlated strongly (Pearson’s r, r2 = 0.75, p < 0.0001; Supplementary Fig. 1).
UltraSEEK Lung Panel analysis
Molecular profiling of ccfDNA was performed using the UltraSEEK® Lung Panel v2 (Agena Bioscience, San Diego, CA, USA) according to the manufacturer’s recommendations22, which comprises analysis of 78 SNVs and indels in BRAF, EGFR, ERBB2, KRAS, and PIK3CA (Supplementary Data 1). 35 μL of the eluate was used in each reaction irrespective of ccfDNA concentration. Mutation calling was performed with the Somatic Variant Reporter using the following analysis parameters: Peak intensity type ‘Area’ with minimum peak intensity of 5 and a minimum z-score of 7. Reported variants were evaluated individually for calling accuracy to prevent false-positive results; the data analyst was blinded for tumor tissue-based molecular outcome.
Tumor specimen handling and tissue NGS
Presence of molecular alterations in the genes for which NSCLC should be currently tested (ALK, BRAF, EGFR, ERBB2, KRAS, MET, NRG1, NTRK1, NTRK2, NTRK3, RET, and ROS1)6 was retrieved from pathology reports. Other reported variants (e.g., mutations in KIT) were recorded as well. In case a variant was identified in plasma that was not reported in the tumor tissue pathology report, the original NGS data was reevaluated for the presence of that specific variant. Tumor tissue specimens were obtained and processed according to standardized protocols25,28. The reported NGS data were generated following routine diagnostic procedures. Tumor tissue samples were subjected to mutational profiling using various NGS approaches (i.e., IonPGM-v0129 and IonPGM-v02b29 hotspot panels on the IonTorrent platform (Thermo Fisher Scientific, Waltham, MA, USA), or smMIP PATHv2D29 and TSO50030 panels on the NextSeq 550 System (Illumina, San Diego, CA, USA)), gene fusion analysis (using either ALK immunohistochemistry (IHC)31, fluorescent in-situ hybridization (FISH)) or RNA analyzes (NanoString transcript analysis (NanoString Technologies, Seattle, WA, USA) or RNA-NGS (Archer DX, Boulder, CO, USA)).
Statistics and Reproducibility
Descriptive statistics were used for patient and tumor characteristics. Agreement between UltraSEEK analysis and tumor tissue NGS was expressed as concordance. For statistical assessment of multiple groups, the Kruskal-Wallis test was performed, followed by Dunn’s multiple comparison test. A binary logistic regression model with backward selection has been constructed to determine whether demographic or clinical (i.e., sex, age, stage of disease, histology, tumor cell percentage, mutated gene, and tumor tissue variant allele frequency [VAF]) and technical (i.e., ccfDNA concentration) parameters affect the mutant detection rate in plasma. GraphPad Prism 8.4.2 software and SPSS statistics version 28 were used for statistical analyzes. A P-value < 0.05 was considered significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Results
Cohort description
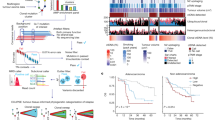
In total, 180 consecutively recruited patients were considered eligible for this study, of whom the majority were diagnosed with lung adenocarcinoma (72%) or lung squamous cell carcinoma (24%; Table 1). Practically all patients had advanced disease (stage IIIB or higher; 98%). Routine diagnostic tumor tissue NGS data were available for 132 NSCLC patients (73%), and fusion gene analysis was performed for 125 (69%; Table 1). Ninety-five pathogenic variants were reported in the patient cohort where at least tumor tissue NGS had been performed (Fig. 1a), of which ten variants and fifteen fusions were not covered by the UltraSEEK Lung Panel (Figs. 2, 3). Forty-six tumors (33%) did not harbor aberrations in the twelve genomic regions of interest Fig. 1a). In the same patient cohort (n = 132), UltraSEEK analysis on plasma-derived ccfDNA identified 56 variants across BRAF, EGFR, KRAS and PIK3CA (Fig. 1a). A combined analysis of tumor tissue and plasma resulted in the detection of at least one variant in 91 patients (69%; Fig. 1c). In plasma, a median VAF of 0.82% (range: 0.11–10.07%) was observed (for all values, see Supplementary Data 2). For each individual patient, clinical data and molecular results are summarized in Fig. 2 and Fig. 3 and detailed in Supplementary Data 2.
a Distribution of variants detected during routine tumor tissue next-generation sequencing (NGS) and that were listed in the pathology report for 132 patients. b Distribution of variants identified using the UltraSEEK Lung Panel on plasma-derived circulating cell-free DNA (ccfDNA) of the same patient cohort. c Distribution of all variants when combining tumor tissue and plasma analysis.
Heatmap representing clinical features (i.e., 1. sex, 2. histology, 3. stage of disease) and molecular analysis (i.e., 4. tumor tissue next-generation sequencing (NGS), 5. tumor tissue fusion and rearrangement analysis, plasma-derived circulating cell-free DNA (ccfDNA) analysis using the UltraSEEK Lung Panel). For variants that can be detected in plasma with UltraSEEK (6. green columns), it was indicated whether this variant was identified in tumor tissue only (orange), plasma only (blue), or in both tumor tissue and in plasma (orange and blue). Variants that were not covered by the UltraSEEK Lung Panel (6. blue columns), as well as fusions and exon skipping (6. orange columns) were displayed separately. For fusions, the fusion partner gene is indicated in the box. If no fusion partner gene was specified, the fusion gene itself was stated in the box. For MET variants, exon 14 indicated that a mutation resulted in skipping of exon 14. Genes that were not analyzed in tumor tissue were highlighted in grey. Based on the molecular profile, eligibility for targeted treatment is indicated (column 7). *G12_G13delinsCA. IHC, immunohistochemistry; FISH, fluorescence in situ hybridization.
Heatmap representing clinical features (i.e., 1. sex, 2. histology, 3. stage of disease) and molecular analysis (i.e., 4. tumor tissue next-generation sequencing (NGS), 5. tumor tissue fusion and rearrangement analysis, plasma-derived circulating cell-free DNA (ccfDNA) analysis using the UltraSEEK Lung Panel). For variants that can be detected in plasma with UltraSEEK (6. green columns), it was indicated whether this variant was identified in tumor tissue only (orange), plasma only (blue), or in both tumor tissue and in plasma (orange and blue). Variants that were not covered by the UltraSEEK Lung Panel (6. blue columns) as well as fusions and exon skipping (6. orange columns) were displayed separately. For fusions, the fusion partner gene is indicated in the box. If no fusion partner gene was specified, the fusion gene itself was stated in the box. For MET variants, exon 14 indicated that a mutation resulted in skipping of exon 14. Genes that were not analyzed in tumor tissue were highlighted in grey. Based on the molecular profile, eligibility for targeted treatment is indicated (column 7). IHC, immunohistochemistry; FISH, fluorescence in situ hybridization.
Concordance with tumor tissue NGS
Concordance between the baseline plasma UltraSEEK Lung Panel analysis and matched tumor tissue NGS was determined considering all variants in the twelve genomic regions of interest that were covered by both panels (i.e., variants in BRAF, EGFR, KRAS) as well as PIK3CA E542, E545, and H1047 mutations (Figs. 2, 3). At patient level, an 82% concordance was observed for variants that could be detected with the tissue- and plasma-based methods (Fig. 4a). More variants were reported with tumor tissue NGS in nineteen patients (14%), while in four patients (3%) more variants were found in plasma. One mutation (KRAS c.34_38delinsTGTGC; p.(G12_G13delinsCA)) that was reported in the tumor tissue affected the same genomic position in plasma but with a different annotation (KRAS c.34 G > T; p.(G12C)), most probably due to the inability of the UltraSEEK Lung Panel to differentiate between covered mutations and certain complex variants at similar coding DNA sequences (Supplementary Data 3). When considering all variants detectable with both methods, a variant-level concordance of 68% was observed (Fig. 4b), which was irrespective of tumor stage (stage III 63%; stage IV 69%).
a Pie chart summarizing the patient-level variant detection rates between tumor tissue next-generation sequencing (NGS) and UltraSEEK Lung Panel analysis on plasma-derived circulating cell-free DNA (ccfDNA). A patient-level concordance (similar variant detection rate in tumor tissue and plasma) of 82% was determined [BRAF, 64% (9/14); EGFR, 100% (9/9); KRAS, 68% (30/44)]. Four mutations [2 in EGFR and 2 in KRAS] were only detected in plasma; one mutation in KRAS incorrectly genotyped in plasma. b Bar graph illustrating variant detection rates in tumor tissue, plasma, or both separately. A variant-level concordance of 68% was observed. All variants and subsequent interpretation are listed in Supplementary Data 2 and Supplementary Data 3.
To determine whether certain confounders affected tumor variant detection in plasma, binary logistic regression analysis with backward selection was performed. The model revealed that ccfDNA input (lower or higher than 20 ng) and mutated gene (either a mutation in BRAF, EGFR or KRAS) affected the variant detection rate in plasma and increased the chance of a false negative result (Supplementary Table 1). No statistically significant results were found when adding only one confounder to the model. This implies that ccfDNA input only affected the detection rate within a gene and not in general. In line, no relation between ccfDNA input and ctDNA detection was observed in a multiple comparison analysis (Supplementary Fig. 2a). However, when stratified per gene (i.e., the detection rate in BRAF, EGFR and KRAS individually), lower median ccfDNA input levels were observed for variants in KRAS not retrieved in plasma (15.5 ng versus 22.5 ng, Mann–Whitney U test, p = 0.027; Supplementary Fig. 2b). For BRAF, the differences in ccfDNA for detected and missed mutations were not significant (37.3 ng versus 29.8 ng, Mann–Whitney U test, p = 0.80; Supplementary Fig. 2b).
Added value of plasma testing in absence of tumor tissue specimens
For 48 patients, tumor tissue NGS data were unavailable (Supplementary Data 4). Insufficient material or poor DNA quality impeded NGS analysis of eleven adenocarcinoma patients (10%). Molecular tumor profiling was not performed for 37 squamous cell carcinoma patients and one pleomorphic carcinoma patient in line with current Dutch guidelines6. A KRAS p.(G12C) mutation was detected inthe plasma of three adenocarcinoma patients and one pleomorphic carcinoma patient. A BRAF p.(V600E) mutation was detected in the plasma of one squamous cell carcinoma patient. Pathogenic PIK3CA mutations were identified in the plasma of three squamous cell carcinoma patients.
Targetable mutation detection and effect on treatment decision making
To assess the potential impact of molecular tumor profiling on therapeutic interventions, theoretical treatment decisions were made in retrospect based on the combined results of tumor tissue and plasma molecular profiling in accordance with the current NCCN guidelines (Figs. 2, 3). Sixty patients (33%) were considered eligible for targeted therapy based on tumor tissue analysis, of which 45 (25%) concerned actionable mutations in BRAF, EGFR and KRAS (Fig. 5a). For 23% of patients, targeted treatments could have been indicated based on mutations identified in plasma (Fig. 5b). When both tumor tissue and plasma analyzes were performed, a total of 67 patients (37%) would have been considered eligible for targeted therapy (Fig. 5c). As such, combined analyzes of tumor tissue and plasma-derived ctDNA identified more patients with actionable targets than tumor tissue testing alone (37% versus 33%, respectively).
Discussion
Liquid biopsy approaches, particularly the detection of genetic aberrations in blood-derived ctDNA, show promising clinical applicability for personalized treatment management16. In absence of a tumor tissue biopsy for predictive testing, cell-free plasma is an alternative source for comprehensive molecular analyzes of lung adenocarcinoma biomarkers11,13. Even when tumor tissue biopsies are available, ctDNA testing may reduce costs and accelerate time-to-treatment12. Although guidelines mandate comprehensive molecular tumor profiling of lung adenocarcinoma, the number of actionable targets remains limited. Therefore, mid-sized targeted panels encompassing the most frequently occurring actionable mutations might meet current clinical demands, as they generally have short hands-on and turnaround times.
Here, the UltraSEEK Lung Panel on the MassARRAY System was used to identify actionable mutations in BRAF, EGFR, ERBB2, and KRAS for NSCLC. Previous studies revealed high concordances between UltraSEEK and other ccfDNA-based mutation detection methods such as ddPCR and the FDA-approved cobas EGFR Mutation Test v222,27. An 82% patient-level concordance was observed for variants that could be detected both with NGS on tumor tissue and the UltraSEEK Lung Panel on plasma. Similar concordances in selected cohorts were reported for using various ctDNA analysis methodologies, including ddPCR (58–69%)25,32, small targeted panels (51–91%)32,33, and broad NGS panels (80–90%)12,19,28,34,35,36. Sixty-eight percent of mutations were concordantly detected between tumor tissue NGS and plasma UltraSEEK analysis, in agreement with previous comparisons22,23,24. Possible explanations for discordances include spatial heterogeneity (intra- and inter-tumor), low to nonexistent shedding of ctDNA, or lack of analytical sensitivity13,37.
Accurate identification of current actionable mutations is of critical importance to guide therapy management. The detection rate of therapeutically targetable mutations in an unbiased cohort was determined in the current study. Similar gene variant distributions (Fig. 1a) were observed as reported previously, as well as the absence of detectable genetic aberrations in 37% of the cases2,4. KRAS p.(G12C) mutations were detected in 20% of lung adenocarcinomas, consistent with the previously reported prevalence in the Netherlands38. A comparable proportion of patients were considered eligible for BRAFV600-, EGFR-, or KRASG12C-targeted therapies based on tumor tissue NGS (25%) or ctDNA analysis (23%), indicating that plasma is a valuable source for tumor mutation analysis. Eight potentially actionable mutations were detected in plasma only, emphasizing the benefit of mutation analysis using liquid biopsy approaches16. Similar studies evaluating baseline plasma samples using ccfDNA-based NGS panels detected actionable variants in 31-36% of NSCLC patients; 25-27% of patients harbored mutations in BRAF, EGFR and KRAS39,40. In the current retrospective study, no clinical decisions were made based on the ctDNA results; therefore, the treatment response and survival outcomes of plasma-indicated therapy could not be assessed. A previous study evaluating the response to plasma NGS-based treatment decisions reported that 86% of patients achieved a durable response to targeted therapy35. Although data on plasma-based treatment guidance is limited, these results suggest that actionable mutations detected in ctDNA could be valuable for the personalized care of patients with advanced NSCLC. Prospective studies are required to ascertain the actual clinical benefit of ctDNA-based molecular profiling.
The limitation of ctDNA-based targeted gene panels – such as the UltraSEEK Lung Panel – is its inability to detect gene fusions, which were present in 8% of patients in our cohort based on tumor tissue. As such, when (actionable) driver mutations cannot be identified in plasma, tumor tissue must be analyzed for the presence of fusion genes. Cell-free plasma-based fusion detection assays are in development and revealed first promising results41,42, however remain far from clinical implementation. Another constraint of the current panel is the inability to determine the plasma tumor fraction. Some plasma-based comprehensive genomic profiling (CGP)-NGS panels (e.g., FoundationOne Liquid CDx, Guardant360 CDx) and shallow whole genome sequencing (sWGS) approaches (e.g., DELFI-tumor fraction) can assess the plasma tumor fraction, which can be regarded as a reliable estimate for tumor burden43.
The data imply that analysis of tumor-derived mutations in plasma with a mid-sized targeted panel such as the UltraSEEK Lung Panel can assist in treatment decision making for NSCLC patients. For UltraSEEK, the average time-to-result – from ccfDNA extraction to variant reporting – was on average sixteen and a half hours (Supplementary Table 2). However, development of a standardized, clinically manageable liquid biopsy workflow is required prior to implementation in routine diagnostics37. In the current workflow, the maximum achievable ccfDNA input from available plasma was analyzed to reach the optimal sensitivity. Previous studies showed higher variant detection rates when using higher ccfDNA inputs using UltraSEEK27. In line, lower ccfDNA inputs impaired the detection rate for variants in BRAF, EGFR, and KRAS in this study (Supplementary Table 1; Supplementary Fig. 2). Increasing plasma volumes result in a higher amount of ccfDNA that can be extracted, but different extraction methods have limitations with respect to the volume of plasma that can be used26.
When no mutations are identified, a comprehensive mutation analysis on tumor tissue is required, as well as for some specific variants with co-occurring (resistance) mutations. For example, recent data suggest that EGFR-mutant NSCLC with concurrent aberrations in TP53 was associated with faster resistance evolution and reduced efficacy of (first-line) osimertinib treatment44,45, which is therefore discouraged in Dutch guidelines45. Furthermore, lung tumors harboring KRAS p.(G12D), STK11 or KEAP1 mutations have poorer treatment outcome to immune therapy28,46,47. Since the number of therapeutically relevant targets is rising, application of more comprehensive liquid biopsy approaches (e.g., AVENIO ctDNA Expanded28, FoundationOne Liquid CDx36, Guardant360 CDx48) could be considered16. Plasma-based NGS assays can evaluate SNVs, indels, CNVs, and chromosomal rearrangements simultaneously; however are more costly and at present time not widely available17. Furthermore, these methods in general demand technical expertise and have longer turnaround times compared to targeted panels20,36. Overall, ccfDNA analyses can be implemented at different stages in the diagnostic process and provide an alternative when molecular analysis of tumor tissue is not possible or reduce time-to-treatment. International guidelines state that genomic profiling on tumor tissue for treatment decision making has to be performed within two weeks after biopsy, which in previous evaluations does not occur in 81% and 93% of cases for KRAS and EGFR testing, respectively49. Comprehensive genomic profiling (CGP) analysis on ctDNA for actionable target evaluation can reduce the time-to-result with approximately two weeks12,50, leading to shorter time-to-treatment as well39. The clinical utility of such a diagnostic strategy must be validated in prospective studies on the clinical implications of ctDNA testing.
The results of ctDNA analyses without knowledge of the histological subtype of the tumor (e.g., small cell lung cancer [SCLC], NSCLC adenocarcinoma or squamous cell carcinoma) may potentially result in overstated conclusions about options for effective targeted therapy. For instance, previous studies question whether EGFR-TKI and ALK inhibitors induce clinical benefit for patients with lung squamous cell carcinoma50. The effectiveness of targeted strategies against BRAF p.(V600E) and KRAS p.(G12C) mutations in lung squamous cell carcinoma is not yet available. Similarly, it remains elusive if activating PIK3CA mutations, which are associated with reduced survival of lung cancer patients51 can also be inhibited with a specific inhibitor as in breast cancer52. As such, interpretation of ctDNA results with respect to treatment options needs to be performed with caution.
In conclusion, combined analyzes of tumor tissue and plasma-derived ctDNA identified more patients with actionable targets than tumor tissue testing alone (37% versus 33%, respectively). A liquid biopsy approach singly might have diagnostic benefit as a pre-screening tool for actionable mutations to reduce time-to-treatment, and as an alternative test when tumor tissue NGS data is unavailable to assist in treatment decision making. Assessment of performance and cost-effectiveness against comprehensive sequencing panels should be performed to determine the diagnostic and clinical value of mid-sized targeted mutation panels.
Data availability
Source data are provided in Supplementary Data 2. Other data from the corresponding author will be made available on reasonable request.
References
Herbst, R. S., Morgensztern, D. & Boshoff, C. The biology and management of non-small cell lung cancer. Nature 553, 446–454 (2018).
Steeghs, E. M. P. et al. Mutation-tailored treatment selection in non-small cell lung cancer patients in daily clinical practice. Lung Cancer 167, 87–97 (2022).
Hendriks, L. E. et al. Oncogene-addicted metastatic non-small-cell lung cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. 34, 339–357 (2023).
Tan, A. C. & Tan, D. S. W. Targeted therapies for lung cancer patients with oncogenic driver molecular alterations. J. Clin. Oncol. 40, 611–625 (2022).
Nationial Comprehensive Cancer Network. Non-Small Cell Lung Cancer. https://www.nccn.org/professionals/physician_gls/PDF/nscl.pdf (2024).
Federatie Medisch Specialisten. Niet kleincellig longcarcinoom. https://richtlijnendatabase.nl/richtlijn/niet_kleincellig_longcarcinoom/startpagina_-_niet-kleincellig_longcarcinoom.html (2024).
Schabath, M. B. & Cote, M. L. Cancer progress and priorities: lung cancer. Cancer Epidemiol. Biomark. Prev. 28, 1563–1579 (2019).
Liam, C. K., Mallawathantri, S. & Fong, K. M. Is tissue still the issue in detecting molecular alterations in lung cancer? Respirology 25, 933–943 (2020).
Liam, C. K. et al. The diagnosis of lung cancer in the era of interventional pulmonology. Int J. Tuberc. Lung Dis. 25, 6–15 (2021).
Merker, J. D. et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J. Clin. Oncol. 36, 1631–1641 (2018).
Chan, H. T., Chin, Y. M. & Low, S. K. Circulating tumor DNA-based genomic profiling assays in adult solid tumors for precision oncology: recent advancements and future challenges. Cancers 14, 3275 (2022).
García-Pardo, M. et al. Association of circulating tumor DNA testing before tissue diagnosis with time to treatment among patients with suspected advanced lung cancer: The ACCELERATE Nonrandomized Clinical Trial. JAMA Netw. Open 6, e2325332 (2023).
Heitzer, E. et al. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 20, 71–88 (2019).
Wan, J. C. M. et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat. Rev. Cancer 17, 223–238 (2017).
Alix-Panabières, C. & Pantel, K. Liquid biopsy: from discovery to clinical application. Cancer Discov. 11, 858–873 (2021).
Heitzer, E. et al. Recommendations for a practical implementation of circulating tumor DNA mutation testing in metastatic non-small-cell lung cancer. ESMO Open 7, 100399 (2022).
Rolfo, C. et al. Liquid biopsy for advanced Non-Small Cell Lung Cancer (NSCLC): A Statement Paper from the IASLC. J. Thorac. Oncol. 13, 1248–1268 (2018).
Abbosh, C., Birkbak, N. J. & Swanton, C. Early stage NSCLC — challenges to implementing ctDNA-based screening and MRD detection. Nat. Rev. Clin. Oncol. 15, 577–586 (2018).
Goldberg, S. B. et al. Early assessment of lung cancer immunotherapy response via circulating tumor DNA. Clin. Cancer Res. 24, 1872–1880 (2018).
Kramer, A. et al. A micro-costing framework for circulating tumor DNA testing in Dutch Clinical Practice. J. Mol. Diagn. 25, 36–45 (2023).
Weber, S. et al. Technical evaluation of commercial mutation analysis platforms and reference materials for liquid biopsy profiling. Cancers 12, 1588 (2020).
van der Leest, P. et al. Detection and monitoring of tumor-derived mutations in circulating tumor DNA using the UltraSEEK Lung Panel on the MassARRAY System in metastatic non-small cell lung cancer patients. Int J. Mol. Sci. 24, 13390 (2023).
Kapeleris, J. et al. Prognostic value of integrating circulating tumour cells and cell-free DNA in non-small cell lung cancer. Heliyon 8, e09971 (2022).
Kulasinghe, A. et al. The identification of circulating tumour DNA using MassARRAY technology in non-small-cell lung cancer (NSCLC). Lung Cancer 160, 73–77 (2021).
van der Leest, P. et al. Circulating tumor DNA as a biomarker for monitoring early treatment responses of patients with advanced lung adenocarcinoma receiving immune checkpoint inhibitors. Mol. Oncol. 15, 2910–2922 (2021).
van der Leest, P. et al. Comparison of circulating cell-free DNA extraction methods for downstream analysis in cancer patients. Cancers 12, 1222 (2020).
Lamy, P. J. et al. Mass Spectrometry as a highly sensitive method for specific circulating tumor DNA analysis in NSCLC: A comparison study. Cancers 12, 3002 (2020).
Weber S. et al. Dynamic changes of circulating tumor DNA predict clinical outcome in patients with advanced non–small-cell lung cancer treated with immune checkpoint inhibitors. JCO Precis. Oncol. 1540–1553 (2021).
Universitair Medisch Centrum Groningen Afdeling Pathologie en Medische Biologie. Next-Generation-Sequence mutatieanalyse. https://www.umcg.nl/-/afdeling/pathologie/moleculaire-diagnostiek/next-generation-sequence-mutatieanalyse (2024).
Universitair Medisch Centrum Groningen Afdeling Pathologie en Medische Biologie. TruSight Oncology 500 (TSO500) Breed DNA NGS panel. https://www.palga.nl/media/uploads/pdf/3/7/370_75-trusight-oncology-500-final-9-2-2022.pdf (2024).
van der Wekken, A. J. et al. Dichotomous ALK-IHC is a better predictor for ALK inhibition outcome than traditional ALK-FISH in advanced non-small cell lung cancer. Clin. Cancer Res. 23, 4251–4258 (2017).
Papadimitrakopoulou, V. A. et al. Epidermal growth factor receptor mutation analysis in tissue and plasma from the AURA3 trial: Osimertinib versus platinum-pemetrexed for T790M mutation-positive advanced non-small cell lung cancer. Cancer 126, 373–380 (2020).
Weber, B. et al. Detection of EGFR mutations in plasma and biopsies from non-small cell lung cancer patients by allele-specific PCR assays. BMC Cancer 14, 294 (2014).
Remon, J. et al. Real-world utility of an amplicon-based next-generation sequencing liquid biopsy for broad molecular profiling in patients with advanced non–small-cell lung cancer. JCO Precis. Oncol. 3, 1–14 (2019).
Aggarwal, C. et al. Clinical implications of plasma-based genotyping with the delivery of personalized therapy in metastatic non-small cell lung cancer. JAMA Oncol. 5, 173–180 (2019).
Bayle, A. et al. Liquid versus tissue biopsy for detecting actionable alterations according to the ESMO Scale for Clinical Actionability of molecular Targets in patients with advanced cancer: a study from the French National Center for Precision Medicine (PRISM). Ann. Oncol. 33, 1328–1331 (2022).
van der Leest, P. & Schuuring, E. Critical factors in the analytical workflow of circulating tumor DNA-based molecular profiling. Clin. Chem. 70, 220–232 (2024).
Garcia, B. N. C. et al. Prevalence of KRAS p.(G12C) in stage IV NSCLC patients in the Netherlands; a nation-wide retrospective cohort study. Lung Cancer 167, 1–7 (2022).
Del Re, M. et al. Clinical utility of Next Generation Sequencing of plasma cell-free DNA for the molecular profiling of patients with NSCLC at diagnosis and disease progression. Transl. Oncol. 41, 101869 (2024).
Russo A. et al. Liquid biopsy of lung cancer before pathological diagnosis is associated with shorter time to treatment. JCO Precis. Oncol. 8:e2300535.
Hasegawa, N. et al. Highly sensitive fusion detection using plasma cell-free RNA in non-small-cell lung cancers. Cancer Sci. 112, 4393–4403 (2021).
Giménez-Capitán, A. et al. Detecting ALK, ROS1, and RET fusions and the METΔex14 splicing variant in liquid biopsies of non-small-cell lung cancer patients using RNA-based techniques. Mol. Oncol. 17, 1884–1897 (2023).
van ‘t Erve, I. et al. Cancer treatment monitoring using cell-free DNA fragmentomes. Nat. Commun. 15, 8801 (2024).
Vokes, N. I. et al. Concurrent TP53 mutations facilitate resistance evolution in EGFR-mutant lung Adenocarcinoma. J. Thorac. Oncol. 17, 779–792 (2022).
Zhao, J. et al. Next-generation sequencing based mutation profiling reveals heterogeneity of clinical response and resistance to osimertinib. Lung Cancer 141, 114–118 (2022).
Nederlandse Vereniging van Artsen voor Longziekten en Tuberculose. Niet-kleincellig longcarcinoom (NSCLC). https://www.nvalt.nl/vereniging/belangrijke-documenten/_/Klinische%20Noodzakelijke%20Targets/KNT-lijst%20NSCLC%20versie%202%202024-01-08.pdf (2024).
Liu, C. et al. KRAS-G12D mutation drives immune suppression and the primary resistance of anti-PD-1/PD-L1 immunotherapy in non-small cell lung cancer. Cancer Commun. 42, 828–847 (2022).
Bauml, J. M. et al. Clinical validation of Guardant360 CDx as a blood-based companion diagnostic for sotorasib. Lung Cancer 166, 270–278 (2022).
Lewis, W. E. et al. Efficacy of targeted inhibitors in metastatic lung squamous cell carcinoma with EGFR or ALK alterations. JTO Clin. Res. Rep. 2, 100237 (2021).
Cui. et al. Up-front cell-free DNA next generation sequencing improves target identification in UK first line advanced non-small cell lung cancer (NSCLC) patients. Eur. J. Cancer 171, 44–54 (2022).
Jee, J. et al. Overall survival with circulating tumor DNA-guided therapy in advanced non-small-cell lung cancer. Nat. Med. 28, 2353–2363 (2022).
Rasti, A. R. et al. PIK3CA mutations drive therapeutic resistance in human epidermal growth factor Receptor 2-positive breast cancer. JCO Precis. Oncol. 6, e2100370 (2022).
Acknowledgements
Plasma samples were provided by the OncoLifeS Biobank of the UMCG. This work was funded by Agena Bioscience.
Author information
Authors and Affiliations
Contributions
P.v.d.L, P.R., L.C.v.K., T.J.N.H. and E.S. conceived and designed this project. P.v.d.L, P.R. and N.R. executed the experimental procedures. P.v.d.L, P.R., A.J.v.d.W., H.K., V.D.d.J., G.S. and L.C.v.K. performed the molecular, clinical, and statistical analyses. All authors contributed to the writing and editing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare the following competing interests: Anthonie J. van der Wekken has a leadership role in the oncology section of NVALT, the guideline committee NSCLC and CUP, and the dure geneesmiddelen committee of NVALT and FMS; is a consultant on advisory boards for AstraZeneca, Eli Lilly, Janssen-Cilag, Roche, and Takeda; has performed lectures for AstraZeneca, BMS, Eli Lilly, Pfizer, and Roche; and has received research grants form AstraZeneca, Boehringer-Ingelheim, and Pfizer. Léon C. van Kempen has a leadership role in the EORTC Melanoma Group and the Commission Personalized Medicine – Belgium; is a consultant on advisory boards for Cyclomics, Janssen-Cilag, LOGEX, Merck Serono, Protyon, and Roche; and has received grants and non-financial support from Amgen, AstraZeneca, Bayer, BMS, Eli Lilly, Janssen-Cilag, LOGEX, Lynxcare, Merck Serono, nanoString, Novartis, Pfizer, and Roche. T. Jeroen N. Hiltermann has a leadership role in the CieBOM; is a consultant on advisory boards for BMS, MSD, Pfizer, and Roche; has received research grants from AstraZeneca, BMS, and Roche; and is involved in clinical studies in collaboration with Amgen, AstraZeneca, BMS, GSK, Merck Serono, Novartis, and Roche. Ed Schuuring has a leadership role in the Dutch Society of Pathology, the European Society of Pathology, the European Liquid Biopsy Society, the CieBOD, and the national guideline committee; is a consultant on advisory boards for Agena Bioscience, Amgen, Astellas Pharma, AstraZeneca, Bayer, BMS, CC Diagnostics, Eli Lilly, GSK, Illumina, Janssen-Cilag, Merck Serono, Novartis, Roche, SinnoVision Lab, and Sysmex; has performed lectures for Agena Bioscience, Biocartis, Bio-Rad, Eli Lilly, Illumina, Roche, and SeraCare Life Sciences; and has received grants and non-financial support from Abbott, Agena Bioscience, ArcherDX, AstraZeneca, Bayer, Biocartis, Bio-Rad, Boehringer Ingelheim, CC Diagnostics, Eli Lilly, Illumina Merck Serono, Roche, and SeraCare Life Sciences. Other authors declare no competing interests.
Peer review
Peer review information
Communications Medicine thanks Fangdi Sun and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. [Peer reviewer reports are available].
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
van der Leest, P., Rozendal, P., Rifaela, N. et al. Detection of actionable mutations in circulating tumor DNA for non-small cell lung cancer patients. Commun Med 5, 204 (2025). https://doi.org/10.1038/s43856-025-00921-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43856-025-00921-8