Abstract
Background
Polyploidy is frequently observed in cancer cells and is closely associated with chromosomal instability, which can lead to cancer progression. Polyploid cancers are more aggressive than diploid cancers, and polyploidy has been shown to be a prognostic marker for hepatocellular carcinoma (HCC). However, polyploidy is challenging to diagnose. Currently, no clinically implementable methods are available for diagnosing polyploidy in cancer.
Methods
We established a method for assessing polyploidization in HCC using deep-learning-based artificial intelligence image recognition models to assess hematoxylin and eosin-stained pathological images. Using 44 HCCs whose ploidy status had been determined by chromosome fluorescence in situ hybridization, we evaluated the ability of our constructed deep learning models to detect HCC ploidy. We then tested the models on an independent group of 169 liver cancers and applied them to a publicly available dataset.
Results
Here we show that our constructed models effectively assess HCC ploidy in a separate cohort and identify a subset with poor prognosis based on the ploidy determinations for 169 HCCs. Our pipeline also identifies HCCs with poor prognosis in the external dataset, with a more significant difference than that for ploidy inferences by genomic analysis. By exploiting the high processing capacity of artificial intelligence, new aspects of polyploid HCC, such as the high prevalence of scirrhous structures, are identified.
Conclusions
Our findings suggest that ploidy assessment using artificial intelligence-based pathological image recognition can serve as a novel diagnostic tool for personalized medicine.
Plain language summary
Cancer cells often have extra sets of chromosomes, a condition called polyploidy. Although polyploidy can be a prognostic marker for liver cancer, there is currently no simple way to detect it in clinical settings. In this study, we developed an artificial intelligence (AI) system that can analyze standard pathology images to assess polyploidy in liver cancer. Our AI accurately identified cases with poorer outcomes based on their polyploidy status, both in our own data and in public datasets. This approach could offer a practical and accessible tool to help doctors better understand each patient’s cancer and support more personalized treatment decisions.
Similar content being viewed by others
Introduction
Hepatocellular carcinoma (HCC), the main type of primary liver cancer, is caused by chronic liver injury, including viral hepatitis and steatotic liver disease, and is one of the most common causes of cancer-related mortality worldwide. HCC often recurs after surgical resection1, and the postoperative 5-year survival rate is less than 60%2. Therefore, identifying HCC patients with poor prognosis who require careful follow-up is a crucial step toward achieving personalized medicine for HCC.
Cancer cell polyploidization is frequently detected in various types of cancers, including HCC3,4. Evaluation of cancer genomes using single nucleotide polymorphism (SNP) arrays and inference of cancer ploidy using the ABSOLUTE algorithm5 has shown that approximately 36% of HCCs exhibit polyploidization3,4. We previously demonstrated that 38% of HCCs are polyploid by directly evaluating chromosome duplications in formalin-fixed paraffin-embedded (FFPE) tissues using fluorescence in situ hybridization (FISH)6. Polyploidization is thought to drive cancer evolution and affect tumor features via various mechanisms, including enhancement of genomic instability7. Polyploid HCC exhibits an aggressive phenotype, and its prognosis is significantly poorer than that of diploid HCC6,8. Polyploid HCC is also likely to exhibit a characteristic pathological appearance, such as macrotrabecular-massive (MTM) histology and occasional involvement of cancer cells with giant nuclei, indicating that polyploidy defines a distinctive subset of HCC6. The diagnosis of polyploidy in HCC would help identify aggressive HCC cases and allow optimization of clinical treatment for HCC.
Unfortunately, the determination of cancer ploidy for HCC has been difficult in routine practice because it requires complicated procedures, such as genomic analysis, chromosome FISH, or image/flow cytometric analysis. Importantly, polyploidy can affect the sizes of tumor cells and nuclei9,10 in addition to the pathological features of HCC, such as the MTM structure6. Considering this, ploidy could potentially be recognized based on morphological features in pathological images. Moreover, recent remarkable advances in artificial intelligence (AI)-based deep learning have enabled the estimation of genetic characteristics of cancers using pathological images11,12. Thus, AI-based examination of hematoxylin and eosin (HE)-stained images, which are routinely obtained in pathological practice, is a promising strategy for identifying polyploid HCC.
In this study, we have developed deep-learning models for diagnosing polyploid HCC by applying image recognition to pathological images. Validations using our separate cohort and the publicly available The Cancer Genome Atlas (TCGA) dataset13 both showed that our model could accurately assess ploidy status in HCC. More importantly, our model identified approximately one-third of HCCs with poor prognosis within our 169-patient cohort and the publicly available TCGA dataset, suggesting that ploidy assessment using our model can meaningfully contribute to HCC prognosis prediction. The diagnosis of polyploid HCC using AI-based evaluation of pathological images is feasible in routine medical practice, even for many patients. This approach is a novel strategy for identifying aggressive HCCs, which enables personalized medicine through treatment optimization.
Methods
Clinical samples and data
A total of 248 HCC tissue samples were obtained from patients who underwent hepatectomy for HCC between 2017 and 2022 at Kobe University Hospital, Kobe, Japan. Of these, 35 cases were excluded because of non-curative resection, perioperative death, or insufficient tissue samples. The remaining 213 cases were divided into datasets and analyzed (Supplementary Fig. 1). FFPE samples were used for hematoxylin and eosin (HE) staining, immunostaining, and multicolored chromosome FISH. Clinical data, including age, sex, and etiology of liver damage, were obtained from the patient’s medical records. This study was performed in accordance with the Declaration of Helsinki and approved by the ethics committee of the Kobe University School of Medicine. Opt-out consent was obtained owing to the retrospective nature of the study.
HE staining and immunostaining
FFPE tissues were sliced into 5 µm-thick sections and subjected to hematoxylin (Sakura 3G, Sakura Finetek, Japan) and eosin (1% Eosin Y solution, Muto Pure Chemicals, Tokyo, Japan) or immunostaining. HE-stained slides were digitized as whole slide images using a Research Slide Scanner VS200 (EVIDENT, Tokyo, Japan) with a ×40 objective lens and subsequently analyzed using OLyVIA v4.1 software (EVIDENT, Tokyo, Japan). Immunohistochemistry was performed using primary antibodies against UBE2C (1:500, ab252940, Abcam) and secondary antibodies, including anti-rabbit (MP-7401, Vector Laboratories, Burlingame, CA, USA) and anti-mouse (MP-7402, Vector Laboratories) antibodies. The immunohistochemically stained slides were photographed under a BX53 light microscope (EVIDENT, Tokyo, Japan).
Chromosome FISH and identification of polyploid HCCs
FFPE samples of 5 µm thickness were stained by multicolored chromosome FISH, using a previously published method6. Briefly, FISH was performed using chromosome enumeration DNA FISH probes targeting chromosomes 7 (#6J36-77), 11 (#6J54-21), and 16 (#5J10-26) following the manufacturer’s instructions (Abbott Laboratories Ltd., IL). The slides were denatured at 72 °C for 6 min, hybridized overnight at 37 °C, and counterstained with DAPI. FISH slides were examined at 100× magnification under a fluorescence microscope (BZ-X710, Keyence). The predominant number of signals for the three stained chromosomes in the tumor nuclei was determined to be the tumor chromosome copy number. To assess HCC ploidy, we calculated the average copy number of the three chromosomes.
Image processing
Three or more regions of interest (ROIs) that exhibited an appropriate appearance without excessive bleeding or necrosis were selected from each whole-slide image. Each ROI had an area of approximately 4 mm2 and was divided into 2048 × 2048-pixel squares. We obtained an average of 251 image tiles for each case, each measuring 2048 × 2048 pixels, and used them for ploidy diagnosis (Supplementary Fig. 2). The length per pixel in these images was 133.477 nm, ensuring sufficient resolution for the analysis.
Deep learning
For deep learning-based classification of tumor ploidy, we adopted multi-instance learning to accommodate a large 2048 × 2048 pixel image size (Supplementary Fig. 3). Each 2048 × 2048-pixel image was further divided into small patches of 256 × 256 pixels, and 42,240 patches were input into a first-stage image classification model such as a conventional neural network (CNN) or Vision Transformer (ViT)14. Among the 42,240 patches, 25,024 and 17,216 were labeled as diploid and polyploid, respectively, according to the chromosome FISH results. The outputs of the patch-level model were aggregated and input into a second-stage model. This approach allowed us to extract local information from the image in the first stage and aggregate information from the entire image in the second stage.
In the first-stage image classification, ImageNet-pre-trained CNN models, including DenseNet12115, ResNet50d16, and EfficientNet_B017 were utilized. All ImageNet-pretrained models used in our study were obtained from the external source, the timm library. Specifically, we used ResNet50 from https://huggingface.co/timm/resnet50.a1_in1k, DenseNet from https://huggingface.co/timm/densenet121.tv_in1k, and EfficientNet from https://huggingface.co/timm/tf_efficientnet_b0.ns_jft_in1k. Their outputs immediately before the last global average pooling layer were extracted, and the extracted features from all patch images were concatenated and input into the global average pooling layer, followed by a linear layer using a method known as PANDA concat tile pooling18. To develop ViT-based models in the first-stage14, we used two self-supervised and pre-trained ViT models. One was pre-trained on our original images Dataset 1 through self-distillation with no labels (DINO)19, a type of self-supervised learning. The other was an encoder of the publicly available hierarchical image pyramid transformer (HIPT)20, which was pre-trained on patch images created from whole slide images in TCGA with DINO. We used a self-supervised pre-trained model to extract HCC-specific information from histopathological images. A learning rate of 0.0005 and a weight decay of 0.04 with the AdamW optimizer21 were used to train our original images with DINO. Training was conducted for 300 epochs, and data augmentation was performed according to the original DINO parameters. We used the ViT-Small model for all the ViT models. The outputs from the last four layers of the ViT were concatenated and input into two-layered gated recurrent unit networks and a linear layer. This approach aims to improve the performance by utilizing the outputs from multiple layers of ViT.
The entire model was trained in two end-to-end stages for 10 epochs using an AdamW optimizer with a learning rate of 0.001 and a weight decay of 0.00005. During end-to-end training, we focused on both the local patch-level information and the global context of the target. All layers were set to be trainable in the CNN-based models, whereas the weights of the first 10 or 11 layers were frozen in the ViT-based models to retain the knowledge acquired through self-supervised learning. We named the model with 11 fixed weights and only the remaining layer trainable as ‘unfrozen1,’ and the model with 10 fixed weights and two remaining layers trainable was called ‘unfrozen2.’ ‘HIPT_unfrozen2’ thus refers to a model that utilizes a self-supervised pre-trained ViT initially trained on publicly available TCGA image data using HIPT. In this model, the weights of the first ten layers were fixed, and the remaining two layers were trainable. In one study, the influence of staining differences between facilities was mitigated by converting the input images to a gray scale. Grayscale conversion was performed using OpenCV’s cv2.COLOR_RGB2GRAY implementation (https://ieeexplore.ieee.org/document/6240859). The model constructed in this experiment is referred to as EfficientNetB0_ Gray.
The performance of the model was validated by dividing the dataset consisting of 44 cases collected at Kobe University Hospital (Dataset 1) into five folds. The ploidy status of these 44 cases was determined using chromosomal FISH. In each iteration, one fold was designated for validation, and the remaining folds were used for training. This process was repeated five times for each single experiment. The predictions for the test data were generated using the average outputs of the five trained models. The cutoff values were determined based on the Youden index22 from the ROC curve in the analysis of Dataset1 and were applied in all analyses.
A custom-built computer server with a CPU (EPYC 7543, Advanced Micro Devices, Inc., Santa Clara, CA, USA) and GPU (A100, 80 GB, NVIDIA Corporation, Santa Clara, CA, USA) was used for all calculations. The operating system used was Ubuntu 20.04 LTS. The implementation of all deep learning models utilized Python 3.9 and PyTorch 1.12.
Evaluation in a large test cohort
A separate cohort of 169 patients (Dataset 3), which was obtained at Kobe University Hospital but not used to construct AI models, was used to validate the AI analysis. Among these, the ploidy status of 38 cases (Dataset 2), a subset of Dataset 3, was determined by FISH. For each case, approximately 280 tiles of 2048 × 2048 pixel images were analyzed using our trained AI models to assess the polyploidization probability of the tumor within each image. The average of these probabilities was calculated as the overall probability of polyploidization in each case. ROC curve analysis using overall polyploidization probability and color map analysis using image-level polyploidization probability were performed using R software (version 4.1.2, R Foundation for Statistical Computing).
TCGA dataset analyses
Whole-slide images of HE-stained FFPE tissues, which are publicly available in TCGA datasets13 (Dataset 4), were analyzed. The dataset consisted of 350 HCC cases whose whole-genome duplication and aneuploidy status had been previously analyzed using the SNP Array5. Hematoxylin and eosin (HE)-stained images and clinical data, including age, sex, tumor stage, grade, vascular invasion, and background liver fibrosis, are also archived in the TCGA database. When setting rectangular ROIs, we attempted to include a sufficiently wide area with well-defined tumor tissue images. However, in 69 cases, more than one-quarter of the ROIs contained features that were unsuitable for pathological tumor diagnosis, such as necrosis, severe fibrosis, and contamination with non-tumor components. These cases were categorized as unsuitable. We also conducted an analysis that excluded them.
Dimensionality Reduction (t-SNE) Analysis
The 2048 × 2048-pixel input images were analyzed using the HIPT_unfrozen2 model, and the embedding vectors for each image immediately before the final linear layer were compressed into two dimensions using t-distributed stochastic neighbor embedding (t-SNE)23 (random_state = 2023, perplexity = 10).
Statistics and reproducibility
Statistical analyses were performed using the Mann–Whitney U test, Student’s t-test, chi-square test, Pearson’s correlation test, or Kaplan–Meier survival analysis using the R software (version 4.1.2, R Foundation for Statistical Computing) and Microsoft Office Excel (Microsoft).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Results
Development of an AI-based image recognition model to estimate HCC ploidy
First, we constructed a model to evaluate the ploidy status of HCC using deep learning and CNN-based image classification. A total of 44 cases whose ploidy status had been determined by chromosome FISH in our previous study6 were used as the training data. The training set included 27 diploid and 17 polyploid HCC cases. After obtaining a whole-slide image of the HE-stained slide for each tumor, we selected three or more ROIs showing the representative pathological appearance of the tumor (Fig. 1a). Each ROI was divided into 2048×2048-pixel tiles, and the tiles were subdivided into 256 × 256 patches for input into the deep-learning algorithm (see Materials and Methods). Deep learning for tumor ploidy classification was performed by training 42,240 small-patch images. The models calculated the probability of tumor polyploidization in each 2048 × 2048-pixel tile, and the average value across all tiles for each tumor was defined as the polyploidy score for the tumor (Fig. 1b, Supplementary Fig. 4).
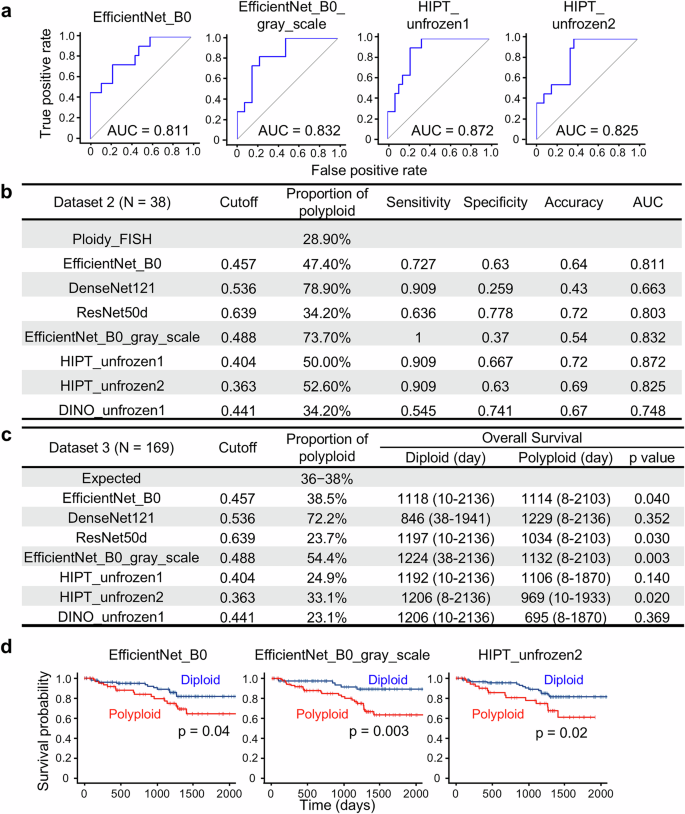
a Scheme for the construction of AI-based image recognition models for determining HCC ploidy. b Representative HE-stained images of ROIs in diploid and polyploid HCC. The probabilities of HCC polyploidization in the corresponding 2048 × 2048-pixel tiles are shown in a color map. Scale bar, 200 μm. c ROC curves and AUC values of representative AI models in cross-validation. The data for the other models are shown in Supplementary Fig. 5. d Evaluation and comparison of the constructed AI models.
We first used three CNN-based image classification models: DenseNet12115, ResNet50d16, and EfficientNet_B017. The validity of the models was assessed by analyzing their receiver operating characteristic (ROC) curves and areas under the curve (AUC)24 (Fig. 1c, Supplementary Fig. 5). Five-fold cross-validation revealed that all three models achieved high AUC values (0.998−1.0, Fig. 1d). For example, with the optimized cutoff value (0.457) of polyploidy score determined based on the ROC curve, the EfficientNet_B0-based model exhibited high accuracy, sensitivity, and specificity (0.977, 1.0, and 0.963, respectively, Fig. 1d, Supplementary Fig. 6). These findings indicate that CNN-based models can be used to evaluate the HCC ploidy status using pathological HE images.
AI-based image recognition successfully assessed HCC ploidy at a low calculation cost
The coloration of HE staining is known to vary due to factors such as fixation conditions and staining protocols, potentially affecting AI model performance25,26. To address this, we constructed a model using EfficientNetB0 on grayscale-converted images to minimize the impact of such variability. The constructed EficientNetB0_gray model showed a high AUC value (0.998), comparable to that of the original CNN-based models, suggesting that the cellular morphological information obtained from grayscale images was sufficient to evaluate HCC ploidy (Fig. 1c, d, Supplementary Fig. 6).
We also developed models using ViT-based architectures, which incur lower calculation costs than CNN-based image recognition. Two and one models were constructed using HIPT20 and DINO19, respectively, both of which enabled the scalability of ViT to large images via self-supervised learning (see Materials and Methods). These encoders were trained on TCGA or liver pathology images obtained at our institution. By freezing parts of the model during training, overfitting can be moderated, even when the labeled data are insufficient. In particular, because it allows for easy replacement of the first stage with other publicly available models trained on pathology images using self-supervised learning, model construction using HIPT requires a shorter learning time than CNN-based models. All three models exhibited high accuracy and AUC values that were comparable to those of the CNN models (Fig. 1d, Supplementary Fig. 6).
AI-based ploidy assessment identified polyploid HCC cases with poor prognosis within a large cohort
We examined whether our constructed AI models could properly assess HCC ploidy using a separate dataset. Tumor ploidy was determined using chromosome FISH in 38 new HCCs (Dataset 2) that were not included in the first dataset (Dataset 1). Their polyploidy scores were then calculated by analyzing their HE images using AI models. The sensitivity, specificity, and proportion of polyploid HCC were determined based on the cutoff values determined in the analysis of Dataset 1 (Fig. 1d, Supplementary Fig. 6). Among the models examined, some, including the two HIPT-based models, exhibited relatively high AUC values over 0.8 (Fig. 2a, b). The decrease in accuracy observed in Dataset 2 compared to Dataset 1 may be attributed to the fact that cases with typical histology of diploid and polyploid cancers were used for training in Dataset 1, while cases in Dataset 2 were selected in an unbiased manner.
a, b Performance of AI models in the validation assessments. The ploidy statuses of 38 HCCs (determined by chromosome FISH) were compared with the ploidy statuses, as assessed by AI models. ROC curves of the representative AI models are shown in (a). c Prognostic stratification based on ploidy assessments by the AI models. A total of 169 HCCs were analyzed. d Kaplan–Meier curves of overall survival. Statistical difference was determined by log-rank test. The three AI models that identified a significant difference in prognosis between diploid and polyploid HCCs in (c) are shown.
To further evaluate the utility of AI-based polyploid HCC identification, a large cohort of 169 HCC cases (Dataset 3) was examined using AI models (Fig. 2c). In particular, the EfficientNet_B0-based and HIPT_unfrozen2 models diagnosed a number of polyploid HCC cases proportional to their prevalence, as shown in previous reports (36–38% 3,6). By identifying polyploidy in HCC, the EfficientNet_B0-based and HIPT_unfrozen2 models discriminated HCC patients with significantly worse overall survival after surgery (Fig. 2c, d, Supplementary Fig. 7). These findings indicate that AI models, especially the HIPT_unfrozen2 model, are useful for identifying polyploid HCC and predicting poor prognosis.
Analysis of a large cohort revealed the characteristics of polyploid HCC
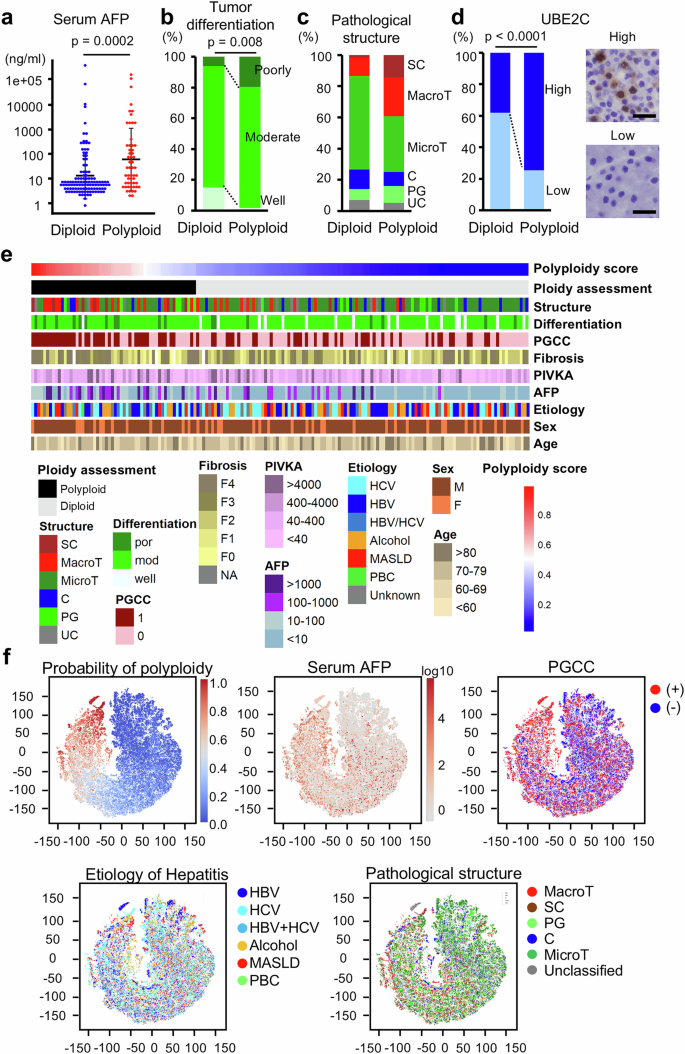
The HIPT_unfrozen2 model, which exhibited the most optimal features for ploidy determination among the constructed models, was used to investigate the characteristics of polyploid HCC by analyzing a large cohort. In Dataset 3, consisting of 169 cases, 113 and 56 cases were diagnosed as diploid and polyploid HCC, respectively, using the HIPT_unfrozen2 model. As observed in other datasets, where no associations were found between tumor ploidy and age, sex, or body mass index (Supplementary Table 1), the two groups showed no significant differences in these variables (Table 1, Supplementary Fig. 8). Consistent with our previous results, serum alpha-fetoprotein (AFP) levels were significantly higher in polyploid HCC than in diploid HCC, whereas tumor size and stage were comparable between the two groups (Table 1, Fig. 3a). Polyploid HCC was also significantly associated with a high prevalence of poor differentiation and exhibited MTM or scirrhous structures (Table 1, Fig. 3b, c). Polyploid giant cancer cells (PGCCs), which exhibit a distinct appearance with prominently large nuclei or profound multinucleation, are frequently observed in polyploid HCC (Table 1). Furthermore, the expression of UBE2C, which we previously reported as a marker suggestive of polyploid HCC, was significantly elevated in polyploid HCC relative to levels in diploid HCC (Fig. 3d). These findings confirm the characteristics of polyploid HCC demonstrated in our previous study and suggest accurate ploidy evaluation by our HIPT_unfrozen2 model. In addition, most polyploid HCCs diagnosed using the AI model did not exhibit well-defined pathological features characteristic of polyploid HCC (Fig. 3e), indicating that the AI model comprehensively assessed ploidy in HCC, considering a complex array of histological information beyond mere tumor structures and differentiation status.
a Serum AFP levels. Error bars indicate mean ± SD. b, c Pathological classification of HCC differentiation and structure. d Immunostaining of UBE2C. Scale Bar 50μm. e Heatmap indicating ploidy scores assessed using the HIPT_unfrozen2 model and clinicopathological features. f t-SNE plots of tile images. Probabilities of polyploidy assessed using the HIPT_unfrozen2 model and clinicopathological features of the tumors are shown. A total of 169 HCCs were analyzed. SC scirrhous, MacroT macro-trabecular, MicroT micro-trabecular, C compact, PG pseudo-glandular, UC unclassified, PIVKA protein induced by vitamin K absence or antagonist II, HBV hepatitis b virus, HCV hepatitis c virus, MASLD metabolic dysfunction associated steatotic liver disease, PBC primary biliary cholangitis.
To further explore the characteristics of polyploid HCC, we visualized case-by-case correlations between the polyploidy scores and clinicopathological features (Fig. 3e). In addition, data derived from all 2048 × 2048-pixel tile images of the 169 HCCs were compressed into two dimensions and visualized using t-SNE plots (Fig. 3f). These plots validated that high serum AFP levels were correlated with high polyploidy probability values calculated using our AI models. Interestingly, HCCs with high polyploidy scores were predominantly positive for PGCCs, highlighting their importance in inferring HCC polyploidy (Fig. 3e). In contrast, hepatitis etiology seemed to exert little influence on HCC ploidy, and HCCs with high polyploidy scores developed in livers with viral hepatitis and steatotic liver diseases (Fig. 3e, f). Our investigation of poorly understood features of polyploid HCC in a large cohort, utilizing the high-throughput analysis capabilities of AI models, verified recently revealed characteristics and provided additional insights.
The AI model robustly identified polyploid HCC in a public dataset and predicted a poor prognosis
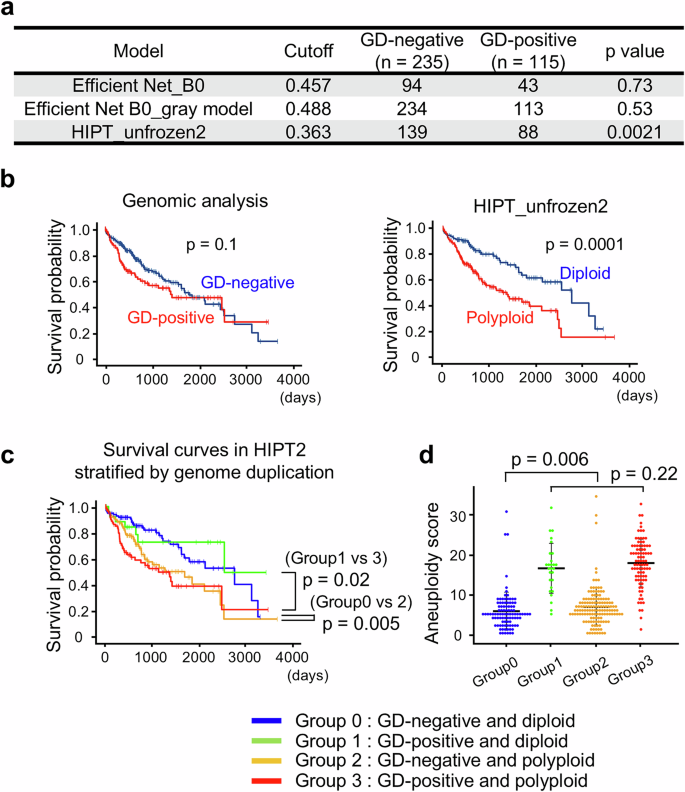
To further verify the utility of the AI-based ploidy discrimination models, we analyzed the HE images of 350 HCC cases in the public TCGA dataset using our representative models, EfficientNet_B0, EfficientNet_B0_gray, and HIPT_unfrozen2. Ploidy assessments obtained by these AI models were compared with a prior determination of genome duplication (GD) by SNP array analysis of tumor genomes4,5. Assessment using the HIPT_unfrozen2 model showed a strong correlation with the GD status determined by genomic analysis (Fig. 4a). The other two models did not demonstrate a significant correlation. Using the GD status based on genomic analysis as a reference, the sensitivity and specificity of the HIPT_unfrozen2 model were 0.77 and 0.41, respectively. Similar to Dataset 3, the polyploid HCC in the TCGA dataset identified by the HIPT_unfrozen2 model showed a high prevalence of PGCC and elevated AFP serum levels, supporting the idea that the AI model can robustly evaluate HCC ploidy status from pathological images obtained under heterogeneous conditions at various facilities (Table 2).
a Conformity between GD detected by genomic analysis and the ploidy status assessed using our AI models. b, c Kaplan–Meier curves displaying overall survival. Statistical difference was determined by log-rank test. d Aneuploidy score. A total of 350 HCC cases in TCGA dataset were divided by their GD status detected by genomic analysis and their ploidy status assessed using the HIPT_unfrozen2 model. Error bars indicate mean ± SD.
We further examined whether the HIPT_unfrozen2 model was helpful in identifying a subset of HCC with poor prognosis. As expected, GD-positive HCC evaluated by genomic analysis showed a trend toward poor prognosis compared to GD-negative HCC, although the difference was weak and insignificant (Fig. 4b). In notable contrast, polyploid HCC identified by the HIPT_unfrozen2 model exhibited markedly poorer prognosis than their diploid counterpart (Fig. 4b). Among the 350 HCCs, the images of 188 cases were designated suboptimal for diagnosis because a substantial proportion of their ROIs were affected by necrosis, severe fibrosis, and contamination with nontumor components. Importantly, however, the HIPT_unfrozen2 model similarly distinguished prognostic differences depending on ploidy status, regardless of the inclusion of these 188 suboptimal cases, highlighting the robust diagnostic capacity of the AI model (Supplementary Fig. 9).
To explore the reasons for the differences in ploidy-related prognostic prediction capability between the HIPT_unfrozen2 model and genomic analysis, TCGA cases were categorized into four groups based on the AI (diploid or polyploid) and genomic results (GD-positive or GD-negative). As expected, GD-positive polyploid HCC had a significantly poorer prognosis than GD-negative diploid HCC (Fig. 4c). Interestingly, polyploid but GD-negative HCC exhibited a poor prognosis, comparable to that of GD-positive polyploid HCC. In addition, diploid but GD-positive HCC showed a good prognosis, similar to that of GD-negative diploid HCC. The HIPT_unfrozen2 model consistently identified HCC with a significantly poorer prognosis regardless of the SNP array results, leading to its superior prognostic prediction over genomic analysis (Fig. 4c). Moreover, among the GD-negative HCC identified using the SNP array, AI-diagnosed polyploid HCC had significantly more chromosomal aberrations than their diploid counterparts (Fig. 4d), suggesting that the AI model distinguished HCC with a poor prognosis by detecting chromosomal instability and polyploidy from pathological images. These findings indicate that our AI model interpreting HCC ploidy status from pathological images can robustly identify HCC with poor prognosis across diverse conditions in multiple facilities.
The HIPT_unfrozen2 model outperforms conventional methods for estimating HCC ploidy from pathological images
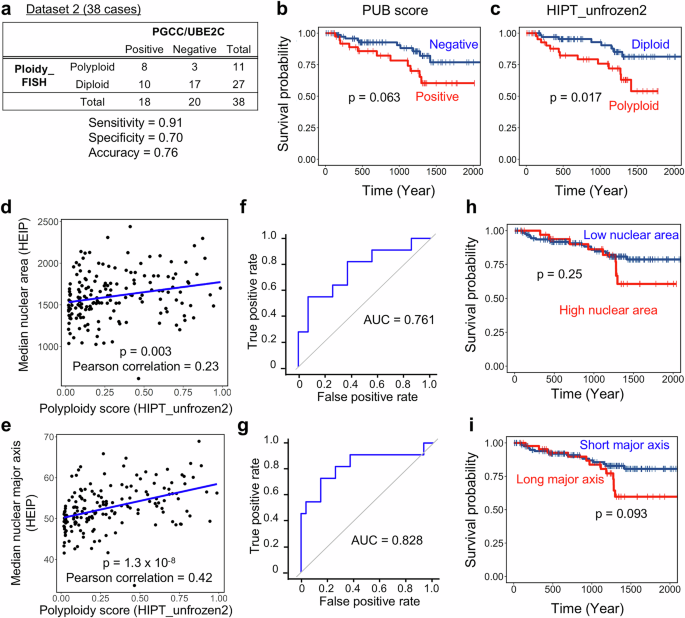
Finally, we compared HIPT_unfrozen2 with existing methods for estimating HCC ploidy from pathological images, evaluating their performance in ploidy classification and prognosis prediction. In our previous study, we proposed a scoring system (PUB score) that combines PGCC detection in HE-stained sections with immunostaining for UBE2C to infer polyploidization in HCC6. When tumors exhibiting both PGCC presence and UBE2C overexpression were classified as polyploid, the PUB classification achieved an accuracy of 0.76 (sensitivity: 0.91, specificity: 0.70) in Dataset 2 (Fig. 5a), which is comparable to that of the AI models. Among the 118 cases in Dataset 3 with available UBE2C immunostaining, the PUB classification identified a group with a poor prognosis, although the difference was not statistically significant (p = 0.063, Fig. 5b). In contrast, HIPT_unfrozen2 distinguished the poor prognosis group more clearly and significantly in the same cases, suggesting that while the combination of PGCC and UBE2C is a useful marker, AI-based ploidy assessment is more effective for predicting prognosis through tumor ploidy classification (p = 0.017, Fig.5c).
a Performance of PUB classification for assessing HCC ploidy. Tumors exhibiting both PGCC presence and UBE2C overexpression were classified as PUB-positive. b, c Kaplan–Meier curves for overall survival. A subset of Dataset 3 (n = 118) with available UBE2C immunostaining was analyzed according to PUB classification and HIPT_unfrozen2 assessment. Correlation between nuclear morphology features extracted by HEIP and the polyploidy score calculated by HIPT_unfrozen2. Median nuclear area (d) and median nuclear major axis (e) were derived from 169 cases in Dataset 3. ROC curves and AUC values for assessing HCC ploidy using median nuclear area (f) or median nuclear major axis (g) extracted by HEIP. Dataset 2 was used for analysis. Kaplan–Meier curves for overall survival analyzed based on the median nuclear area (h) or median nuclear major axis (i). Cases in Dataset 3 were stratified using cutoff values determined by ROC curves in f, g based on the Youden method. In b, c, f, g, statistical significance was assessed using the log-rank test.
We also compared HIPT_unfrozen2 with another published AI-based tool that assesses tumor ploidy by evaluating nuclear morphology, the HE Image Processing pipeline (HEIP)27, using the same HE-stained images analyzed in our study. After segmenting cell nuclei, we identified tumor nuclei using the HEIP algorithm and assessed tumor ploidy based on two morphological features: nuclear area, which is known to correlate with ploidy28, and the nuclear major axis, which was reported as the most strongly correlated feature in the original study27. As expected, both the median tumor nuclear area and the median nuclear major axis extracted by HEIP showed a highly significant correlation with the polyploidy score calculated by HIPT_unfrozen2, suggesting that HEIP accurately captured tumor nuclear morphology (Fig. 5d, e). Using Dataset 2, where tumor ploidy was confirmed by chromosome FISH, we assessed the performance of HEIP in tumor ploidy classification through ROC analysis, yielding AUC values comparable to that of HIPT_unfrozen2 (0.761 for nuclear area and 0.828 for the nuclear major axis, Fig. 5f, g). We further examined the prognostic utility of HEIP-based tumor ploidy assessment in Dataset 3. When tumors were stratified by the nuclear area, no significant difference in prognosis was observed between the high (n = 35) and low (n = 134) groups (log-rank, p = 0.25, Fig. 5h). Stratification using the nuclear major axis showed better separation of prognostic groups, but the difference remained statistically insignificant (log-rank, p = 0.093, Fig. 5i).
Taken together, these findings indicate that HIPT_unfrozen2 outperforms conventional methods in classifying tumor ploidy and stratifying prognosis based on pathological images of HCC.
Discussion
Polyploidy is a hallmark of the human cancer genome and a crucial cause of chromosomal instability (CIN), predisposing cells to chromosomal aberrations. As CIN promotes cancer heterogeneity and clonal evolution, cancers exhibiting polyploidy and CIN have a poor prognosis regardless of the organ of origin, as shown by recent pan-cancer analyses29,30. Assessment of polyploidy can thus help evaluate the risk of cancer evolution and predict tumor prognosis. In this study, we developed a method that can rapidly and efficiently assess HCC ploidy and predict poor prognosis utilizing AI-based recognition of HE pathological image features.
Previous studies exploring the impact of polyploidy in cancer have often been based on genomic analysis methods, including SNP arrays and whole-genome sequencing, which are expensive, labor-intensive, and time-consuming. Moreover, inferring tumor ploidy from genomic data is challenging and can sometimes be inaccurate owing to factors such as tumor purity and intratumor heterogeneity31,32. Contamination by diploid non-tumor cells is also unavoidable in flow cytometric analysis, making accurate evaluation of tumor ploidy challenging. In contrast, chromosome FISH, which was used to obtain a reference label in this study, enables direct assessment of chromosome numbers in individual tumor cells. However, its labor-intensive procedures limit its scalability, making it difficult to conduct high-throughput assessments on large sample sets or wide tissue regions. In addition, genomic analysis, flow cytometry, and FISH all require expensive and specialized technologies, presenting considerable barriers to their routine use in clinical practice. In contrast to these methods, our AI-based pathological image recognition approach provides a simple and cost-effective method for tumor ploidy evaluation, requiring only HE-stained images. It also enhances throughput and enables ploidy assessment across large tumor regions while capturing intratumoral heterogeneity, which was previously unattainable. Furthermore, unlike genomic analysis and flow cytometry, our technology preserves pathological information and enables integration with additional analyses to study tumor architecture and the microenvironment. Combining it with spatial transcriptomics and emerging techniques will further enhance our understanding of tumor ploidy.
Among the advantages of our AI-based ploidy assessment pipeline, one of the key features is its capacity to process large areas at high resolution. In our model, ploidy status spanning wide tumor areas could be determined for each small 280 μm square area, corresponding to a 2048 × 2048-pixel tile. Furthermore, the results were comprehensively assessed with the visualization of intratumoral heterogeneity. As cancers exhibit diverse cellular features within a single tumor, the above-mentioned advantages would contribute to accurate tumor ploidy assessment and prognostic prediction. Another advantage of our AI model is that it can comprehensively determine HCC ploidy using single-cell morphology information, tissue architecture, and accompanying nontumor cells. We successfully developed such AI models, capable of utilizing diverse histological information, by adopting a balanced approach, selectively fine-tuning encoder layers while leveraging self-supervised pretraining. In contrast to conventional deep-learning methods, where encoders and aggregation layers are trained either jointly or separately, our approach expanded the trainable parameters for learning complex ploidy features while reducing overfitting and preserving valuable representations. Consequently, our model, which incorporates an integrative pathological assessment, was more effective for prognosis prediction than conventional image-based methods that rely solely on nuclear morphology (Fig. 5)27. While ploidy estimation relying on nuclear morphology remains valuable for studies focusing on single-cell characteristics of tumor cells, our model will be particularly useful in clinical settings where determining treatment strategies based on tumor properties is essential. Since pathologists still cannot infer HCC ploidy from HE-stained images alone, our AI model provides a pathway to integrating ploidy assessment into clinical practice.
These advantages may have permitted more precise identification of HCC with poor prognosis using our AI model than that provided by the SNP array-based ploidy assessment in the TCGA dataset. Notably, the HIPT_unfrozen2 model distinguished HCCs with a higher aneuploidy score and poorer prognosis among those classified as GD-negative by the SNP array. The aneuploidy score, reflecting numerical chromosomal abnormalities, is often used as a surrogate for CIN, as tracking chromosomal changes over time or measuring their diversity is impractical in bulk genomic analyses. However, complementary abnormalities within a tumor may be averaged out, and CIN does not always correlate with the aneuploidy score. Like tumor ploidy, CIN is often difficult to assess from bulk genomic data. It remains unclear whether tumors with an elevated aneuploidy score (Group 2 in Fig. 4) were actually polyploid because of inaccuracies in genomic data analysis or were near-diploid cancers that had lost chromosomes from a polyploid state33,34,35. However, given the close relationship between polyploidy and CIN, and their shared importance in predicting cancer prognosis and guiding treatment strategies, the HIPT_unfrozen2 model, which efficiently evaluates ploidy-related tumor aggressiveness and identifies HCC with poor prognosis by incorporating pathological information, is precious for clinical management of HCC. Further investigation is needed to determine whether the HIPT_unfrozen2 model specifically focuses on a subgroup of polyploid HCC with poorer prognosis.
Analysis of a large number of HCCs utilizing the high processing ability of AI confirmed previous findings about some features of polyploid HCC, including the MTM structure and UBE2C overexpression6, and unveiled some new aspects of polyploid HCC in the present study. Among the pathological structures of HCC, the scirrhous type was markedly more prevalent in polyploid HCC than in diploid HCC, although it was relatively rare compared to the MTM structure. This suggests that tumor cell ploidy may significantly influence spatial tumor architecture and the microenvironment. As polyploidization may involve humoral factor secretion36, further investigation into its microenvironmental impact is warranted. Moreover, our AI model detected polyploid HCC arising from various etiologies of liver diseases. Since the hepatic zones and genomic alterations affected by liver injury vary depending on the etiology, our findings suggest that the carcinogenic mechanisms and origins of polyploid HCC can stem from a variety of genomic abnormalities and locations.
The application of AI-based pathological image analysis for tumor ploidy assessment has the potential to revolutionize clinical practice. Diagnosis of tumor ploidy using our technology allows for the rapid evaluation of tumor ploidy in parallel with histopathological diagnosis from HE-stained tissue. AI-based tumor ploidy assessment, completing a 4 mm2 region in just 0.06 s on our computational setup, is remarkably faster than other ploidy evaluation methods. This speed allows tumor ploidy to be used as a prognostic indicator without delaying treatment, enabling personalized strategies like intensive treatments and appropriate follow-ups for patients with poor outcomes. Moreover, clinical research targeting KIF18A, a promising therapeutic target for tumors with polyploidy or chromosomal instability3,37,38,39, is currently underway (NCT05902988). If such treatments become clinically viable, our technology will serve as a companion diagnostic tool for targeted treatments of polyploid cancers.
In conclusion, we have developed a method for assessing HCC ploidy using AI-based image recognition. Similar strategies can be applied to various types of cancer. Given the aggressive features of polyploid and CIN-positive cancers originating in different organs, AI-based assessment of tumor ploidy could contribute to the identification of treatment-resistant cancers with poor prognosis, regardless of the cancer type. Prospective trials using samples with varied processing conditions across multiple institutions will validate the utility of our technology and facilitate clinical application.
Data availability
The numerical data plotted (source data) in the graphs in Figs. 3a, 4d, 5d, e, and Supplementary Fig. 8 are in Supplementary Data 1. The datasets generated and analyzed during the current study are not publicly available because of patient privacy concerns, but are available from the corresponding authors upon reasonable request.
Code availability
The complete source code for both the model and data preprocessing is available at https://github.com/abebe9849/LiverNploid40.
Abbreviations
- AFP:
-
alpha-fetoprotein
- FFPE:
-
formalin-fixed paraffin-embedded
- FISH:
-
fluorescence in situ hybridization
- HCC:
-
hepatocellular carcinoma
- UBE2C:
-
ubiquitin-conjugating enzyme E2
References
Tsilimigras, D. I. et al. Recurrence patterns and outcomes after resection of hepatocellular carcinoma within and beyond the Barcelona Clinic Liver Cancer criteria. Ann. Surg. Oncol. 27, 2321–2331 (2020).
Reveron-Thornton, R. F. et al. Global and regional long-term survival following resection for HCC in the recent decade: A meta-analysis of 110 studies. Hepatol. Commun. 6, 1813–1826 (2022).
Quinton, R. J. et al. Whole-genome doubling confers unique genetic vulnerabilities on tumour cells. Nature 590, 492–497 (2021).
Taylor, A. M. et al. Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell 33, 676–689.e673 (2018).
Carter, S. L. et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 30, 413–421 (2012).
Matsuura, T. et al. Histological diagnosis of polyploidy discriminates an aggressive subset of hepatocellular carcinomas with poor prognosis. Br. J. Cancer 129, 1251–1260 (2023).
Lens, S. M. A. & Medema, R. H. Cytokinesis defects and cancer. Nat. Rev. Cancer 19, 32–45 (2019).
Bou-Nader, M. et al. Polyploidy spectrum: a new marker in HCC classification. Gut 69, 355–364 (2020).
Øvrebø, J. I. & Edgar, B. A. Polyploidy in tissue homeostasis and regeneration. Development 145, dev156034 (2018).
Shu, Z., Row, S. & Deng, W. M. Endoreplication: The good, the bad, and the ugly. Trends Cell Biol. 28, 465–474 (2018).
Kather, J. N. et al. Pan-cancer image-based detection of clinically actionable genetic alterations. Nat. Cancer 1, 789–799 (2020).
Fu, Y. et al. Pan-cancer computational histopathology reveals mutations, tumor composition and prognosis. Nat. Cancer 1, 800–810 (2020).
Cancer Genome Atlas Research Network Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 169, 1327–1341.e1323 (2017).
Zhai, X., Kolesnikov, A., Houlsby, N. & Beyer, L. Scaling Vision Transformers. Preprint at https://doi.org/10.48550/arXiv.2106.04560 (2021).
Huang, G., Liu, Z., van der Maaten, L. & Weinberger, K. Q. Densely Connected Convolutional Networks. Preprint at https://doi.org/10.48550/arXiv.1608.06993 (2016).
He, T. et al. Bag of Tricks for Image Classification with Convolutional Neural Networks. Preprint at https://doi.org/10.48550/arXiv.1812.01187 (2018).
Tan, M. & Le, Q. V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. Preprint at https://doi.org/10.48550/arXiv.1905.11946 (2019).
Bulten, W. et al. Artificial intelligence for diagnosis and Gleason grading of prostate cancer: the PANDA challenge. Nat. Med 28, 154–163 (2022).
Caron, M. et al. Emerging Properties in Self-Supervised Vision Transformers. Preprint at https://doi.org/10.48550/arXiv.2104.14294 (2021).
Chen, R. J. et al. Scaling Vision Transformers to Gigapixel Images via Hierarchical Self-Supervised Learning. Preprint at https://doi.org/10.48550/arXiv.2206.02647 (2022).
Loshchilov, I. & Hutter, F. Decoupled Weight Decay Regularization. Preprint at https://arxiv.org/abs/1711.05101 (2019).
Youden, W. J. Index for rating diagnostic tests. Cancer 3, 32–35 (1950).
van der Maaten, L. & Hinton, G. Visualizing Data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Hanley, J. A. & McNeil, B. J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 143, 29–36 (1982).
Boschman, J. et al. The utility of color normalization for AI-based diagnosis of hematoxylin and eosin-stained pathology images. J. Pathol. 256, 15–24 (2022).
Tellez, D. et al. Quantifying the effects of data augmentation and stain color normalization in convolutional neural networks for computational pathology. Med Image Anal. 58, 101544 (2019).
Ariotta, V. et al. H&E image analysis pipeline for quantifying morphological features. J. Pathol. Inf. 14, 100339 (2023).
Wen, Z. et al. Deep-Learning-Based Hepatic Ploidy Quantification Using H&E Histopathology Images. Genes 14, 921 (2023).
Bielski, C. M. et al. Genome doubling shapes the evolution and prognosis of advanced cancers. Nat. Genet 50, 1189–1195 (2018).
van Dijk, E. et al. Chromosomal copy number heterogeneity predicts survival rates across cancers. Nat. Commun. 12, 3188 (2021).
Luo, Z., Fan, X., Su, Y. & Huang, Y. S. Accurity: accurate tumor purity and ploidy inference from tumor-normal WGS data by jointly modelling somatic copy number alterations and heterozygous germline single-nucleotide-variants. Bioinformatics 34, 2004–2011 (2018).
Zaccaria, S. & Raphael, B. J. Accurate quantification of copy-number aberrations and whole-genome duplications in multi-sample tumor sequencing data. Nat. Commun. 11, 4301 (2020).
Matsumoto, T. et al. Proliferative polyploid cells give rise to tumors via ploidy reduction. Nat. Commun. 12, 646 (2021).
Ben-David, U. & Amon, A. Context is everything: aneuploidy in cancer. Nat. Rev. Genet. 21, 44–62 (2020).
Laughney, A. M., Elizalde, S., Genovese, G. & Bakhoum, S. F. Dynamics of tumor heterogeneity derived from Clonal Karyotypic Evolution. Cell Rep. 12, 809–820 (2015).
Arnandis, T. et al. Oxidative stress in cells with extra centrosomes drives non-cell-autonomous invasion. Dev. Cell 47, 409–424.e409 (2018).
Marquis, C. et al. Chromosomally unstable tumor cells specifically require KIF18A for proliferation. Nat. Commun. 12, 1213 (2021).
Payton, M. et al. Small-molecule inhibition of kinesin KIF18A reveals a mitotic vulnerability enriched in chromosomally unstable cancers. Nat. Cancer 5, 66–84 (2024).
Cohen-Sharir, Y. et al. Aneuploidy renders cancer cells vulnerable to mitotic checkpoint inhibition. Nature 590, 486–491 (2021).
Abe, M. LiverNploid. https://github.com/abebe9849/LiverNploid (2025).
Acknowledgements
The results here are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga. This work was supported by the Japan Agency for Medical Research and Development, AMED under Grant Number JP22fk0210099, Seed Incubation Grant from Osaka University, and a Transdisciplinary Research Project from the Institute for Datability Science, Osaka University.
Author information
Authors and Affiliations
Contributions
ToM designed the study. Ta.M., M.K., Y.U. and Y.K. obtained clinicopathological information, and Ta.M. obtained image data for analysis. Ta.M. and Y.H. performed chromosomal FISH. M.A. and H.N. constructed the AI models. Ta.M., M.A., H.N. and To.M. analyzed the data and wrote the manuscript. M.K., H.N., Y.U., Y.K. and E.H. critically reviewed the manuscript. All authors have approved the final version of the manuscript and agreed to its publication.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communication Medicine thanks Ko Sugawara, Sofia Ferreira, and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Matsuura, T., Abe, M., Harada, Y. et al. Selective identification of polyploid hepatocellular carcinomas with poor prognosis by artificial intelligence-based pathological image recognition. Commun Med 5, 270 (2025). https://doi.org/10.1038/s43856-025-00967-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s43856-025-00967-8