Abstract
Background
Sonographic cervical length is a powerful predictor of maternal risk for spontaneous preterm birth (sPTB). Twin and family studies have established a maternal genetic heritability for sPTB ranging from 13 to 20%, however, there is no corresponding estimate for the heritability of mid-trimester cervical length, or an understanding of how genetic factors contribute to cervical changes across pregnancy.
Methods
This study was based on a prospective longitudinal cohort of (N = 5,160) Black/African American women who underwent serial sonographic examination of the uterine cervix during pregnancy and were genotyped via next-generation low-pass whole genome sequencing.
Results
Bivariate genetic correlations estimated using genome-wide complex trait analysis (GCTA) indicated that a large proportion of the genes influencing cervical change across pregnancy also influenced gestational duration. SNP-level associations were observed near genes involved in the progesterone, estrogen, and insulin signaling pathways.
Conclusions
These results suggest that a large proportion of genetic loci for preterm birth exert their influence through the process of cervical remodeling. Polygenic profiling of maternal genetic liability to cervical shortening could aid in the development of clinical risk assessment tools to identify high-risk women who may benefit from more frequent cervical length screening and earlier interventions to prevent preterm delivery.
PLAIN LANGUAGE SUMMARY
This study looked at how a pregnant woman’s genes may affect the length of her cervix during pregnancy. A short cervix can increase the risk of giving birth too early (preterm birth). Researchers studied over 5000 Black/African American women and found that some of the same genes that affect how long a pregnancy lasts also affect how the cervix changes. These genes are linked to hormone pathways like progesterone and estrogen. Understanding these genetic links could help doctors identify women at higher risk for early delivery and provide care sooner to help prevent it.
Similar content being viewed by others
Introduction
The cervix undergoes carefully orchestrated compositional changes throughout pregnancy, transitioning from a firm structure that protects the in utero environment during pregnancy to a soft and pliable tissue allowing for parturition1,2,3. In most pregnancies delivered at term, a gradual decrease in cervical length (CL) is observed around 30 weeks of gestation, shortening from approximately 35–45 mm to complete effacement by the first stage of labor4. However, this process of cervical shortening begins earlier or happens more rapidly in some pregnancies. A short cervix has been defined in different ways5 but a sonographic cervical length of less than 25 mm before 24 weeks of gestation is a generally accepted definition, and is associated with a 6-fold increase in the risk for preterm birth (i.e., birth before 37 completed weeks)4. The faster cervix shortens, and the earlier in pregnancy that shortening begins, the higher the risk for spontaneous preterm delivery.
There is a clear pathophysiological connection between cervical dysfunction and spontaneous preterm birth, and several maternal risk factors are associated with both conditions, including maternal age, parity, pre-pregnancy BMI, race, and previous abortion6,7,8,9,10,11,12. A recent study showed that many maternal risk factors for preterm birth are mediated through their effects on cervical change during pregnancy13. Yet, whether the previously estimated genetic influences on spontaneous preterm birth14,15,16,17,18 are related to or operate through mechanisms that affect cervical length remains unclear. While genome-wide association studies (GWAS) have identified a handful of loci associated with gestational duration19,20, there have been no reported heritability estimates of cervical length and only limited efforts to identify individual loci associated with cervical dysfunction21.
Meta-analyses of twin and family studies have established that nearly all human complex traits are influenced by genetic factors22. An average heritability of 45.1% was observed across 64 traits in heritability studies specifically related to female reproduction, indicating that nearly half of interindividual variation in these traits could be attributed to genetic factors22,23. Estimates for the heritability of spontaneous preterm birth range from 14 to 40% based on twin and family studies24 and 13–21% based on single nucleotide polymorphism (SNP) genotype data18,19. Heritability estimates are often correlated within functional domains, suggesting that traits related to a particular biological function share similar genetic influences. The extent to which changes in cervical length and gestational duration share a genetic architecture (i.e., the number, location, frequency, interactions, and effect sizes of genetic variants influencing each trait) can be quantified by calculating their bivariate genetic correlation. A better understanding of the genetic and mechanistic relationships between cervical changes during pregnancy and gestational duration is needed to make progress in predicting, preventing, and treating adverse maternal and neonatal outcomes associated with preterm delivery.
Since there is currently no characterization of the genetic architecture of cervical length during pregnancy or how these genetic factors relate to gestational duration, this study focused on the following fundamental questions: What aspects of cervical length change across pregnancy are heritable (i.e., mid-pregnancy length, linear, and nonlinear change components)? To what extent do genetic influences on cervical change reflect a polygenic pattern of inheritance? And how much of this genetic contribution, if any, is shared with genes affecting gestational duration?
Heritability estimates and the genetic correlations among traits were estimated by conducting a GWAS alongside Genome-Wide Complex Trait Analysis (GCTA) of low-pass whole-genome sequencing data in a cohort of pregnant Black/African American women with serial cervical length measurements obtained across pregnancy. Additional insight into the biological mechanisms underlying cervical change during pregnancy was gained by integrating GWAS results with functional and pathway annotations, including tissue-specific gene expression data, epigenetic marks, and chromatin accessibility.
Methods
Cohort description and phenotyping
The genotyped cohort was composed of self-identified Black/African-American women from the Detroit, Michigan area enrolled in a prospective study of pregnant women. Serial sonographic CL measurements between 8 and 40 weeks of gestation were available for 5160 women carrying singleton pregnancies. In this study, gestational duration was evaluated as both gestational age at delivery (GAD) and spontaneous preterm birth (sPTB). GAD was measured in weeks from the first day of a woman’s last menstrual period (confirmed by ultrasound) to the day of delivery. sPTB were included for uninduced deliveries before 37 completed weeks of gestation, including births following both spontaneous preterm labor and preterm premature rupture of membranes (PPROM). Patients treated with therapies expected to alter the natural progression of cervical shortening during pregnancy were excluded. Cervical change across gestation was quantified for each pregnancy in a previous study using Latent Growth Curve Analysis (LGCA)13,25, a cross-disciplinary analytic approach that leverages repeated measurements to estimate how a trait changes within and between individuals over time. Interindividual differences in cervical change during pregnancy were described by coefficients of the growth curve estimated using LGCA methods corresponding to the model intercept, linear change (i.e., slope), and nonlinear (i.e., quadratic) change. The model intercept was centered at 17–20 weeks so that the estimated value of this coefficient would correspond to the midtrimester cervical length (MTCL). The cervical change coefficients and gestational duration measures (i.e., GAD, sPTB) were used as traits in the GCTA and GWAS analyses.
Whole-genome sequencing and quality control
Purified genomic DNA (gDNA) was extracted from buffy coat samples and genotyped by BGI Americas Corporation using paired-end low-pass, whole genome sequencing. Gencove, Inc. performed the sequence alignment using the GRCh37-hg19 human reference genome assembly, variant calling, phasing, and imputation based on the loimpute v0.18 algorithm for low-pass sequencing data26. A recombination map derived from the HapMap II project was used to interpolate recombination rates across all sites in the imputation reference panel. After imputation, genetic variant information was available for 65 million SNPs.
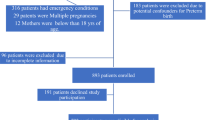
The mean effective sequencing depth coverage of the sample was 1.45x. Variants with a genotyping rate of <5% (geno <0.05); a MAF of <0.5% (MAF < 0.005); or that were found to be in extreme deviation from Hardy-Weinberg equilibrium (HWE < 0.000001) were excluded. Approximately 19.8 million single-nucleotide polymorphisms (SNPs) remained after these quality control filters. A principal component analysis (PCA) of genotypes was conducted in Plink27,28 based on an external reference panel of known ancestry reference (1000 Genomes Project, Phase 3)29. Samples were excluded due to poor quality (n = 2); missing data (n = 15); duplicates (n = 80); and population stratification concerns (n = 243). In total, 4423 samples passed all quality control measures for genetic analyses (Fig. 1).
Purified genomic DNA from 5023 patients with available samples were sent for low-pass whole-genome sequencing. Approximately 9.8 million single-nucleotide polymorphisms (SNPs) remained after variant level filtering for genotyping rate (geno <0.05), minor allele frequency (MAF < 0.01), and deviation from Hardy-Weinberg equilibrium (HWE < 0.000001). Samples from 4423 women remained after excluding samples for poor nucleic acid quality (n = 2), missing data (n = 15), duplicates (n = 80), and population stratification (n = 243).
Trait heritability and bivariate genetic correlation analysis
The SNP-based heritability of cervical change coefficients was estimated using restricted maximum likelihood (REML) as implemented in GCTA version 1.92.330. Covariates in the heritability analysis included sequencing depth, batch number, maternal height, pre-pregnancy BMI, parity, self-reported race/ethnicity, and the first 10 SNP-based ancestry principal components (PCs). Both self-reported race/ethnicity and ancestry principal components were included as covariates as they capture different aspects of environmental and genetic factors that may account for trait variance. SNP-based heritability was estimated for maternal height, pre-pregnancy BMI, GAD, and sPTB to compare the within-cohort estimates to SNP and twin-based heritability published in the literature. Bivariate GCTA was used to decompose observed correlations between traits into covariance components explained by genetic and environmental factors. Bivariate GCTA methods were used to estimate the genetic correlation between the cervical change coefficients and gestational duration measures.
Genome-wide associations
Although identification of an appreciable set of individual SNP associations was not deemed practical given the available sample size31,32, an exploratory analysis of genome-wide associations was conducted to gain insight into potential genetic mechanisms and biological pathways associated with cervical change and pregnancy duration. The whole genome association analysis toolset Plink (PLINK v2.00a3.3LM 64-bit Intel)27,28 was used to test for genetic associations with cervical change coefficients and gestational duration. PLINK dosages for the X chromosome in the all-female samples were set to levels similar to regular diploid chromosomes. Generalized linear models were used to test associations for the quantitative traits (cervical length coefficients, and GAD), while a logistic regression model was used for sPTB. Sequencing depth, batch number, self-reported race/ethnicity, height, pre-pregnancy BMI, parity, and the first 10 PCs from the PCA were variance-standardized and included as covariates. Due to the shorter extent of linkage disequilibrium (LD) in populations with African ancestry, individual SNP associations in this cohort were evaluated using the more conservative threshold for genome-wide significance (p < 1.6 × 10−8) based on a Bonferroni correction for an estimated 3.0 million independent LD blocks.
Annotation of GWAS findings
The integrative web-based platform FUMA33 was used to annotate variant-level summary statistics from suggestive loci (p < 1.0 × 10−5) associated with cervical change coefficients and gestational duration outcomes. Functional annotation and the mapping of SNPs to nearby genes was performed using the SNP2GENE function, while GENE2FUNC was used to obtain insights into putative biological mechanisms of mapped genes. Tissue specific expression patterns for the genes mapped by SNP2GENE were assessed in 53 tissue types using RNA-seq data from the Adult Genotype Tissue Expression (GTEx) v834. Gene-wise associations35 and Multi-marker Analysis of GenoMic Annotation (MAGMA) of the combined set of loci associated with the cervical change coefficients was used for exploratory gene-set enrichment analyses35 for a comprehensive test of genetic effects on cervical length change.
Pre-registration
Analyses were pre-registered with the Center for Open Science using the AsPredicted format (https://osf.io/km2cg).
Study approval
Participants were enrolled under the protocols for Biological Markers of Disease in the Prediction of Preterm Delivery and for Preeclampsia and Intra-Uterine Growth Restriction: A Longitudinal Study (WSU IRB#110605MP2F and NICHD/NIH# OH97-CH-N067) between 2005 and 2017 at the Center for Advanced Obstetrical Care and Research (CAOCR) at Hutzel Women’s Hospital. The Institutional Review Boards of Wayne State University and the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)/National Institutes of Health (NIH)/U.S. Department of Health and Human Services (DHHS) (Detroit, MI, USA) approved the study. All participants provided written informed consent for the collection of cervical length data and blood samples for future genetic research studies.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Results
SNP-based heritability estimates for cervical length change
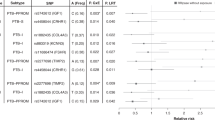
Cervical change during pregnancy was found to be highly heritable in the cohort of 4,423 women retained for genetic analyses (Table 1). Cervical length change trajectories were characterized by estimating the mid-trimester cervical length (i.e., intercept), the linear (i.e., slope), and nonlinear (i.e., quadratic) values for each pregnancy using longitudinal growth curve analysis (LGCA) (Fig. 2). Genetic sources explained at least 50% of inter-individual trait differences in cervical change during pregnancy (h2Intercept = 0.509 [95% CI 0.327, 0.691]; h2Linear = 0.509 [95% CI 0.323, 0.695]; h2Nonlinear = 0.507 [95% CI 0.325, 0.689]) (Fig. 3, Supplementary Table 1). The estimated heritability of GAD (h2GAD = 0.158 [95% CI −0.022, 0.338)] and spontaneous preterm birth (h2sPTB = 0.154 [95% CI −0.024, 0.332)] were consistent with previously reported values obtained by twin and family methods18,19. For instance, the SNP-based heritability estimated for GAD in this cohort was remarkably similar to the only twin and family study16 that has estimated maternal heritability for GAD in African American women (i.e., 15.8% vs. 13.8%). Heritability estimates for height (h2Height = 0.675 [95% CI 0.506, 0.844]) and BMI (h2BMI = 0.576 [95% CI 0.400, 0.752]) were also consistent with published estimates for these traits in other cohorts36,37. These results show that cervical changes during pregnancy are highly heritable, and that heritability estimates from this cohort align with those reported in other studies.
Marginal plots show distribution of CL data across gestational ages and birth outcome classes. Average trajectories of CL change during pregnancy estimated by LGCA are shown for four classes of clinical birth outcomes (extremely preterm births delivered before 28 weeks gestation, very preterm births delivered before 32 weeks gestation, late preterm births delivered before 37 weeks gestation, and full term births delivered after 37 completed weeks of gestation) are shown in color. The trajectory and rate of cervical change during pregnancy varies by gestational age at delivery. Interindividual differences in cervical change during pregnancy were described by coefficients of the growth curve estimated using latent growth curve analysis (LGCA) methods corresponding to the model intercept, linear change (i.e., slope), and nonlinear (i.e., quadratic) change.
Values in the upper triangle represent observed phenotypic correlations between traits, while values in the lower triangle represent genetic correlations between traits. Values along the diagonal represent a trait’s SNP-based heritability. Note that negative genetic correlations occur when an increase in trait 1 corresponds with a decrease in trait 2.
Genetic correlations between cervical length change and gestational duration
Bivariate GCTA analysis of cervical change coefficients and gestational duration suggest that genetic factors significantly contribute to the observed phenotypic correlations between these traits. The observed correlations between the intercept, linear, and nonlinear cervical change coefficients (rIntercept-Linear = 0.850 [95% CI 0.846, 0.854], rIntercept-Nonlinear = −0.658 [95% CI −0.670, −0.646], rLinear-Nonlinear = −0.642 [95% CI −0.654, −0.629]) indicated that a shorter midtrimester cervical length (i.e., intercept) was associated with a more rapid rate of cervical shortening that begins earlier in pregnancy (Fig. 3). There was significant overlap between the genetic factors influencing cervical change coefficients and gestational age at delivery rg, Intercept-GAD = 0.714 [95% CI 0.419, 0.873]; rg, Linear-GAD = 0.982 [95% CI 0.951, 0.993]; rg, Nonlinear-GAD = −0.490 [95% CI −0.766, −0.062] (Fig. 3, Supplementary Table 2). Moreover, the genetic correlation between measures of gestational duration, GAD, and sPTB (rg, GAD-sPTB = −0.600 [95% CI −1.114, −0.086]), was similar to another reported estimate20. These results confirm a genetic correlation between changes in cervical length and gestational age at delivery, suggesting they share underlying biological mechanisms.
Genome-wide associations with gestational duration
An exploratory genome-wide analysis was conducted to identify potential genetic mechanisms associated with gestational duration. Two variants were found to be significantly associated with GAD using a genome-wide significance threshold of 1.6 × 10−8 (Fig. 4D). SNPs rs75296056 and rs185443461 on Chromosome 18 (p = 1.85 × 10−9) are located near the gene encoding the Ras Like Without CAAX 2 (RIT2) protein (Supplementary Fig. 1). RIT2 is a calmodulin-binding GTPase that belongs to the RAS superfamily, and is thought to play a role in cellular signaling, calcium dynamics, dopamine trafficking, and autophagy38,39. While the relationship between RIT2 and gestational duration is not clear, it may promote the maintenance of pregnancy by regulating the autophagy-lysosomal pathway, a cellular process of protein degradation that provides energy and nutrients to maintain cellular homeostasis under stressful environmental conditions39. Autophagic activity is essential to the processes of embryonic development, implantation, and placental development in the hypoxic and low-nutrient intrauterine environment of early pregnancy. Dysregulation of placental autophagy has been linked to several pregnancy complications, including miscarriage, gestational hypertension, gestational obesity, gestational diabetes, intrauterine growth restriction, and preterm birth39,40. In total, 4410 candidate SNPs, 400 independent lead SNPs, 329 genomic risk loci, and 950 mapped genes were identified using a “suggestive” association threshold of 1 × 10 − 5 (Fig. 4D). The QQ plots for the GWAS and gene-based testing (Fig. S2) show an acceptable level of genomic inflation (λSNP = 1.32). These results are consistent with a polygenic architecture, wherein many genes each contribute a small amount to interindividual variation in gestational duration. Genome-wide analysis in this cohort identified two novel loci significantly associated with gestational age at delivery, which were mapped to genes with functions relevant to the maintenance of pregnancy.
Genome-wide associations with cervical change
Exploratory genome-wide analyses were also performed to identify genetic associations with measures of cervical change during pregnancy. While no associations met the estimated threshold for genome-wide significance in populations with African ancestry (p < 1.6 × 10−8), associations with two did surpass the standard Bonferroni correction for multiple testing in GWAS (p < 5.0 × 10−8). Using this threshold, rs148405652 on Chromosome 4 (p = 3.85 × 10−8) showed a strong association with estimated mid-trimester cervical length (i.e., intercept coefficient) (Fig. 4A). This variant maps to the Trypsin-like serine protease gene (TMPRSS11D), a Type II transmembrane serine protease that is involved in regulating many cellular processes relevant to cervical remodeling, including activation of metalloproteases, degradation of extracellular matrix components, fibroblast-to-myofibroblast differentiation, epithelial-mesenchymal transition, microvascular angiogenesis, mobilization of intracellular Ca2+, and activation latent growth factors, cytokines and hormones41,42.
Previously known as human airway trypsin-like protease (HAT), TMPRSS11D/HAT is highly expressed in cervical and vaginal tissue samples and has been detected in the proteome of cervical-vaginal fluid34,43,44. This versatile, membrane-anchored serine protease is linked to a pro-inflammatory immune response and increased mucus production in tissues of the respiratory, lymphoid, and gastrointestinal systems41 and has been shown to modulate the pathogenicity, spread, and tissue-specificity of viral infections45. TMPRSS11D/HAT may therefore play a crucial role in supporting the adaptive immune and innate barrier functions of mucosal surfaces in the female reproductive tract46,47,48.
The midtrimester cervical length estimate was also associated with rs73591716 on Chromosome 10 (p = 4.63 × 10−8) (Fig. 4A). This variant maps to the gene encoding calcium voltage-gated channel auxiliary subunit beta 2 (CACNB2), a subunit of a voltage-dependent calcium channel protein that controls the influx of calcium ions in cardiomyocytes, regulating the rate and contractile force of cardiomyocytes49. This gene has been previously associated with Brugada syndrome, an inherited condition that predisposes patients to fatal cardiac arrhythmias50,51,52, and risk for severe psychiatric disorders53. Calcium ion (Ca2+) signaling plays a fundamental role in regulating uterine myometrial activity during labor and delivery, and calcium channel blockers are used clinically as tocolytic drugs to suppress uterine contractions and delay preterm labor54,55,56,57. Notably, a candidate gene study of in myometrial samples from PPROM patients reported that the expression of several beta subunits of the L-type calcium channel CACNB was significantly higher in patients who delivered preterm compared to full-term deliveries58.
An association was also observed between the linear coefficient and rs139045636 on Chromosome 3 (p = 7.67 × 10−8) which is located within the gene encoding Interleukin 12 A + Antisense RNA (IL-12A-AS1) and near the gene encoding Interleukin 12 A (IL12A) (Fig. 4B). Interleukin-12 is a pro-inflammatory cytokine produced primarily by myeloid cells, such as dendritic cells and macrophages, which induces interferon-γ (IFNγ) expression and promotes T helper 1 (Th1) differentiation, among other functions59,60. During pregnancy, there is a shift from the pro-inflammatory microenvironment associated with pregnancy establishment to a homeostatic state that persists for most of gestation to promote maternal-fetal immune tolerance61. A breakdown of such maternal-fetal tolerance has therefore been associated with adverse pregnancy outcomes, including preterm labor61,62. Molecular studies suggest that IL-12 concentrations in cervical secretions are predominantly the result of local cytokine production63. Interestingly, cervical concentrations of IL-12 have been associated with many maternal risk factors for a short cervix and preterm labor, including age, parity, genital tract infection, and a high vaginal pH64,65,66.
A threshold of 1 × 10−5 was then used to identify “suggestive” SNPs for exploratory analyses. In total, 150 independent genomic loci containing 300 genes were identified at this threshold across all three cervical length coefficients describing cervical change during pregnancy (Fig. 4A–C). Of these, several suggestive SNPs were mapped to genes involved in key hormonal pathways for pregnancy; namely, Low Density Lipoprotein (LDL) Receptor Related Protein 1B (LRP1B) (rs141249340, p = 9.81 × 10−8), Relaxin 2 (RLN2) (rs35460272, p = 1.06 × 10−7), RAS like estrogen regulated growth inhibitor (RERG) (s11056394, p = 7.45 × 10−7), the Progesterone Receptor (PGR) (rs147395300, p = 7.73 × 10−7), Growth Regulating Estrogen Receptor Binding 1 (GREB1) (rs13431344, p = 3.64 × 10−6), and pregnancy-associated plasma protein A (PAPPA) (rs7865534, p = 7.14 × 10−6). Suggestive associations were also identified between cervical change coefficients and several proteins involved in connective tissue organization, including SPARC-Related Modular Calcium-Binding Protein 2 (SMOC2)(rs571424410, p = 5.36 × 10−8), ADAM metallopeptidase with thrombospondin type 1(ADAMTS1)(rs113157601, p = 3.14 × 10−8), Lysyl Oxidase Like 3 (LOXL3) (rs6546907, p = 3.46 × 10−7), Matrix Metallopeptidase 17 (MMP17) (rs74495243, p = 8.92 × 10−6), Fibulin 1 (FBLN1) (rs115293381, p = 9.37 × 10−6), and fibrillar collagens COL3A1 and COL5A2 (rs9776810, p = 9.74 × 10−6).
Functional annotation and gene-set enrichment results
Functional annotations and enrichment analyses were performed on the variants associated with cervical change. Genes mapped to suggestive hits showed downregulated expression in GTEx v8 endocervical tissue (Supplementary Fig. 3). Gene-set enrichment profiling of all genes mapped to suggestive SNPs for cervical change resulted in significant enrichment of molecular and functional domains based on a false discovery rate (FDR) of 5% (Supplementary Tables 4–12). Identified Kegg and Gene Ontology (GO) pathways corresponded to biological processes and molecular functions related to growth factors, energy metabolism, hormonal signaling, immune response, and inflammation (Supplementary Tables 8–13), including the Interleukin-2 pathways (p = 6.56×10−6) and the Interleukin-20 (p = 9.12 × 10−6), Interferon (p = 1.97 × 10−5), and Vascular Endothelial Growth Factor (VEGFA, p = 4.88 × 10−4) signaling pathways. Glucose and lipid metabolism (p = 1.57 × 10−4) are vital for fetal development and energy homeostasis during pregnancy, and alterations in these metabolic pathways could negatively impact fetal growth and development, affecting gestational length67,68. Estrogen signaling (p = 1.91 × 10−4) plays a significant role in preparing the body for labor and delivery by promoting the production of prostaglandins, lipid compounds that mediate cervical ripening and stimulate myometrial contractions to initiate labor69. Likewise, activation of immune cells and the release of cytokines contributes to an inflammatory response in the uterus that is conducive to the onset of labor70,71.
Differentially expressed gene sets from 54 tissue types (GTEx v8) suggested that many of the mapped genes are differentially expressed in endocervical cells (Supplementary Fig. 3). In contrast to the ectocervix, which protrudes into the vagina and is covered by stratified squamous epithelial cells, the endocervix is composed of the columnar epithelial cells that line the cervical canal and internal os and may be influenced by molecular and hormonal signaling from the chorioamniotic membranes69,72,73. Although these results suggest that many of the mapped genes could be expressed in endocervical tissue, the differential regulation results should be interpreted with caution as the 10 cervical tissue samples included in GTEx V8 were collected from non-pregnant donors, many of whom were likely postmenopausal, and thus do not reflect gene expression changes that occur during pregnancy.
Discussion
Proper cervical length remodeling throughout pregnancy is integral to successfully maintaining a pregnancy and preparing for delivery. While there are many suspected factors that can precipitate early labor74,75, pathological shortening and effacement of the cervix during pregnancy provides a clear mechanism of preterm birth4,76. This study was designed to provide the first assessment of the genetic architecture of cervical length and quantify its genetic relationship to gestational duration. Low-pass sequencing experiments allowed for the estimation of the number, effect size, and population frequency of genetic variants essential for heritability studies of cervical length and its rate of change across pregnancy.
The findings of the current study have contributed two fundamental insights. First, genetic factors accounted for approximately 50% of inter-individual differences in cervical change during pregnancy. These heritability estimates are consistent with a highly polygenic, complex trait influenced by both genetic and environmental factors74,75. The estimated heritability of gestational duration outcomes and other biometric measurements were consistent with published estimates for these traits obtained by twin and family and SNP-based estimates in other studies18,19,36,37. Specifically, the estimated SNP-based maternal heritability of GAD from this study of self-identified African American women (h2SNP = 15.8%) was similar to the only estimate available from an African American twin and family cohort (h2Twin = 13.8%)16. The tendency for SNP-based heritability estimates to be lower than their twin-based counterparts77 was not observed and preliminarily suggests that common genetic alleles, in contrast to rare or structural variants, accounted for the majority of genetic variation in gestational age at birth.
Second, the robust genetic correlations estimated among cervical change and gestational duration traits suggested that a large proportion of the genetic factors contributing to cervical change also influenced gestational duration. Estimates of the genetic correlation along with the trait-specific heritability can be used to provide an approximation of the observed bivariate trait correlations expected to be due to correlated genetic factors78. For example, the observed trait correlation between GAD and the MTCL (i.e., intercept) measures was 0.232 of which 87.1% was estimated to be due to shared genetic factors. Similarly, the GAD-linear (rGAD-Linear = 0.246) and GAD-nonlinear (rGAD-Nonlinear = −0.148) trait correlations were estimated to be almost entirely due to genetic factors, near 100% and 93.9%, respectively. A meta-analysis of genetic correlations across human diseases and traits confirms that a genetic correlation (rg) of 0.30 or higher is a reasonable expectation for two traits with a robust epidemiological association, such as cervical length and gestational duration79. A point worth noting was that the replication of the GAD-sPTB genetic correlation in the current study (rg = −0.600 [95% CI −1.114, −0.086]), which was first described by Solé-Navais et al. (rg = −0.62 [95% CI −0.72, −0.51])20, does not confirm the genetic similarity of these traits but rather indicates the loss of genetic information after dichotomizing GAD15.
The gene set enrichment and pathway analysis of GWAS summary statistics have shed light on biological mechanisms involved in cervical changes during pregnancy and gestational duration. Pathway enrichment analysis of suggestive variants in this cohort revealed an increased burden in molecular processes related to hormone and growth factor signaling, energy metabolism, tissue remodeling, inflammation, and immune regulation. These biological pathways have been previously implicated in the onset of labor in both preterm and term birth. Genomic signals near genes involved in insulin, progesterone, and estrogen signaling, and various cytokines and growth factors like IL-2, IL-20, VEGFA, and interferon signaling were also identified. These results align with current knowledge regarding hormonal influences and responses in cervical tissue, and could offer a potential mechanistic explanation for why intervention with vaginal progesterone might help prolong gestation and prevent preterm birth in women with a short cervix.
Several variants were found to be associated with cervical change and gestational duration. RLN2 is an ovarian hormone that promotes loosening of connective tissues in the pelvic ligaments and the cervix to allow for easier passage of the infant during labor and delivery80. Similar to progesterone, relaxin is thought to promote pregnancy maintenance and uterine quiescence to prevent spontaneous abortions and the preterm onset of labor81. Although studies addressing the specific functions of RERG and GREB1 in pregnancy are limited, these proteins are thought to play a role in regulating hormone-responsive pathways in reproductive tissues82,83,84,85. Other hormonally-responsive genes identified herein include LoxL3, which is shown to be physiologically regulated by estrogen in mice86,87,88, and FBLN1, a calcium-binding secreted glycoprotein with repeated epidermal-growth-factor-like domains that is also regulated by estrogen89,90,91.
A number of other genes identified herein are implicated in tissue remodeling processes in the context of pregnancy, including MMP17, a membrane-type matrix metalloproteinase that degrades components of the extracellular matrix during tissue remodeling related to embryonic development and reproduction92. Fibulins are associated with the formation and rebuilding of extracellular matrices and elastic fibers, including connective tissue disorders in humans93, and FBLN1 has been associated with HPV infection leading to cervical precancerous lesions and cervical carcinoma94. COL3A1 and COL5A2 produce alpha chains for a low abundance fibrillar collagens, and mutations in these gene are associated with Ehlers-Danlos syndrome, types I and II, which themselves are associated with an increased risk for cervical insufficiency, sPTB, and PPROM74,95. While its specific function during pregnancy is not understood, LRP1B has been implicated in modulating inflammatory responses and and may interact with extracellular matrix proteins or cell surface receptors to regulate cell adhesion and migration in the uterus and placenta during pregnancy96,97. Finally, PAPPA encodes a metalloproteinase that regulates the Insulin-like Growth Factor (IGF) pathway98, and low serum levels of PAPPA in the first trimester have been associated with an increased risk of many adverse pregnancy outcomes, including preterm birth, low birth weight, preeclampsia, miscarriage, and stillbirth99,100.
The current study was designed to characterize several features of the genetic architecture of cervical change during pregnancy, but was not designed to identify a comprehensive set of common SNP effects. Larger sample sizes will be needed to detect robust statistical associations for individual genetic variants. This study did not address the influence of fetal genes, the vaginal microbiome, or the broader set of environmental and sociodemographic factors that may also contribute to cervical change during pregnancy. Additionally, further study is required to understand the mechanisms behind the generation of genetic correlations between these traits, which could be due to different forms of pleiotropy, linkage disequilibrium, or uncontrolled sources of bias101.
Previous genetic studies of preterm birth have focused on individuals of European ancestry in Scandinavia and the Americas, limiting the generalization of findings across diverse global populations. This project was the first large genomic study of birth outcomes in a cohort of US-based women with African ancestry, who are underrepresented in genomic studies, and suffer disproportionately higher rates of sCL and sPTB. Conducting research in a high-risk and underrepresented population could contribute to more effective, personalized prevention strategies for patients from a similar genetic and sociodemographic background.
Collectively, the data herein provide evidence for a substantial genetic influence on changes in cervical length throughout pregnancy. Moreover, such genetic factors are highly correlated with those influencing pregnancy duration, implying shared causal loci between these traits. By undertaking functional and pathway annotation of the observed genetic variants, multiple individual genes and signaling pathways were identified that may be implicated as determinants of cervical changes and gestational duration. Although genetic correlations have not been widely applied to birth outcomes research, they have been extensively used in the fields of psychiatric genetics and endocrinology/cardiology to predict genetic liability for correlated outcomes, enhance causal and mediation analyses, and increase statistical power for multi-trait genetic association studies79. Thus, establishing the shared genetic risk between cervical shortening and gestational duration may improve the utility of cervical length as a screening tool, provide targets for future pharmacogenetic studies, and encourage broader screening programs to identify women with a short cervix who would benefit from clinical interventions, with the goal of preventing preterm birth.
Data availability
Deposition of whole genome sequencing data in a public repository is prohibited due to ethical and legal constraints however GWAS summary statistics have been uploaded to the NHGRI-EBI GWAS Catalog (GCST90623790, GCST90623791, GCST90623792, GCST90623793). These include the SNP rs number, chromosome position, effect allele, minor allele frequency, test statistic, standard error, and pvalue.
Code availability
Scripts for processing whole genome sequencing data, GWAS, and GCTA analysis are available at the Open Science Framework (https://osf.io/5cngs).
References
Timmons, B., Akins, M. & Mahendroo, M. Cervical remodeling during pregnancy and parturition. Trends Endocrinol. Metab. 21, 353–361 (2010).
Naftolin, F. & Stubblefield, P. G. Dilatation of the Uterine Cervix: Connective Tissue Biology and Clinical Management. (Raven Press, New York, 1980).
Gc, L. Cervical ripening as an inflammatory reaction. The cervix in pregnancy and labor. Clinical and biochemical investigation 1–9 (1981).
Iams, J. D. et al. The Length of the Cervix and the Risk of Spontaneous Premature Delivery. N. Engl. J. Med. 334, 567–573 (1996).
Hassan, S. S. et al. Patients with an ultrasonographic cervical length <or =15 mm have nearly a 50% risk of early spontaneous preterm delivery. Am. J. Obstet. Gynecol. 182, 1458–1467 (2000).
Anum, E. A., Brown, H. L. & Strauss, J. F. 3rd Health disparities in risk for cervical insufficiency. Hum. Reprod. 25, 2894–2900 (2010).
Goldenberg, R. L., Culhane, J. F., Iams, J. D. & Romero, R. Epidemiology and causes of preterm birth. Lancet 371, 75–84 (2008).
Kandil, M., Sanad, Z., Sayyed, T. & Ellakwa, H. Body mass index is linked to cervical length and duration of pregnancy: An observational study in low risk pregnancy. J. Obstet. Gynaecol. 37, 33–37 (2017).
Buck, J. N., Orzechowski, K. M. & Berghella, V. Racial disparities in cervical length for prediction of preterm birth in a low risk population. J. Matern. Fetal Neonatal Med. 30, 1851–1854 (2017).
Cho, S. H. et al. Maternal Characteristics, Short Mid-Trimester Cervical Length, and Preterm Delivery. J. Korean Med. Sci. 32, 488–494 (2017).
Brittain, J. J. et al. Prior Spontaneous or Induced Abortion Is a Risk Factor for Cervical Dysfunction in Pregnant Women: a Systematic Review and Meta-analysis. Reprod. Sci. https://doi.org/10.1007/s43032-023-01170-7.(2023).
Gudicha, D. W. et al. Personalized assessment of cervical length improves prediction of spontaneous preterm birth: a standard and a percentile calculator. Am. J. Obstet. Gynecol. 224, 288.e1–288.e17 (2021).
Wolf, H. M. et al. Risk factors for spontaneous preterm birth are mediated through changes in cervical length. medRxiv https://doi.org/10.1101/2023.04.20.23288082.(2023).
Lunde, A., Melve, K. K., Gjessing, H. K., Skjaerven, R. & Irgens, L. M. Genetic and environmental influences on birth weight, birth length, head circumference, and gestational age by use of population-based parent-offspring data. Am. J. Epidemiol. 165, 734–741 (2007).
York, T. P. et al. Fetal and maternal genes’ influence on gestational age in a quantitative genetic analysis of 244,000 Swedish births. Am. J. Epidemiol. 178, 543–550 (2013).
York, T. P., Strauss, J. F., Neale, M. C. & Eaves, L. J. Racial differences in genetic and environmental risk to preterm birth. PLoS One 5, e12391 (2010).
Svensson, A. C. et al. Maternal effects for preterm birth: a genetic epidemiologic study of 630,000 families. Am. J. Epidemiol. 170, 1365–1372 (2009).
York, T. P., Eaves, L. J., Neale, M. C. & Strauss, J. F. III The contribution of genetic and environmental factors to the duration of pregnancy. Am. J. Obstet. Gynecol. 210, 398–405 (2014).
Zhang, G. et al. Genetic Associations with Gestational Duration and Spontaneous Preterm Birth. N. Engl. J. Med. 377, 1156–1167 (2017).
Solé-Navais, P. et al. Genetic effects on the timing of parturition and links to fetal birth weight. Nat. Genet. 55, 559–567 (2023).
Volozonoka, L. et al. Genetic landscape of preterm birth due to cervical insufficiency: Comprehensive gene analysis and patient next-generation sequencing data interpretation. PLoS One 15, e0230771 (2020).
Claussnitzer, M. et al. A brief history of human disease genetics. Nature 577, 179–189 (2020).
Polderman, T. J. C. et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat. Genet. 47, 702–709 (2015).
Wadon, M., Modi, N., Wong, H. S., Thapar, A. & O’Donovan, M. C. Recent advances in the genetics of preterm birth. Ann. Hum. Genet. 84, 205–213 (2020).
Wolf, H. M. et al. Study protocol to quantify the genetic architecture of sonographic cervical length and its relationship to spontaneous preterm birth. BMJ Open 12, e053631 (2022).
Li, J. H., Mazur, C. A., Berisa, T. & Pickrell, J. K. Low-pass sequencing increases the power of GWAS and decreases measurement error of polygenic risk scores compared to genotyping arrays. Genome Res 31, 529–537 (2021).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015).
1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
Visscher, P. M. et al. 10 Years of GWAS Discovery: Biology, Function, and Translation. Am. J. Hum. Genet. 101, 5–22 (2017).
Wu, T., Liu, Z., Mak, T. S. H. & Sham, P. C. Polygenic power calculator: Statistical power and polygenic prediction accuracy of genome-wide association studies of complex traits. Front. Genet. 13, 989639 (2022).
Watanabe, K., Taskesen, E., van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017).
GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020).
de Leeuw, C. A., Mooij, J. M., Heskes, T. & Posthuma, D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 11, e1004219 (2015).
Yengo, L. et al. A saturated map of common genetic variants associated with human height. Nature 610, 704–712 (2022).
Guo, J. et al. Quantifying genetic heterogeneity between continental populations for human height and body mass index. Sci. Rep. 11, 5240 (2021).
Tebar, F. et al. Pleiotropic roles of calmodulin in the regulation of KRas and Rac1 GTPases: Functional diversity in health and disease. Int. J. Mol. Sci. 21, 3680 (2020).
Obergasteiger, J. et al. The small GTPase Rit2 modulates LRRK2 kinase activity, is required for lysosomal function and protects against alpha-synuclein neuropathology. NPJ Parkinsons Dis. 9, 44 (2023).
Zhou, P., Wang, J., Wang, J. & Liu, X. When autophagy meets placenta development and pregnancy complications. Front Cell Dev. Biol. 12, 1327167 (2024).
Menou, A. et al. Human airway trypsin-like protease, a serine protease involved in respiratory diseases. Am. J. Physiol. Lung Cell. Mol. Physiol. 312, L657–L668 (2017).
Antalis, T. M., Bugge, T. H. & Wu, Q. Membrane-anchored serine proteases in health and disease. Prog. Mol. Biol. Transl. Sci. 99, 1–50 (2011).
Muytjens, C. M. J., Yu, Y. & Diamandis, E. P. Discovery of antimicrobial peptides in cervical-vaginal fluid from healthy nonpregnant women via an integrated proteome and peptidome analysis. Proteomics 17, 1600461 (2017).
Uhlén, M. et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Yamaoka, K. et al. Cloning and characterization of the cDNA for human airway trypsin-like protease. J. Biol. Chem. 273, 11895–11901 (1998).
Sheng, Y. H. & Hasnain, S. Z. Mucus and Mucins: The Underappreciated Host Defence System. Front. Cell. Infect. Microbiol. 12, 856962 (2022).
Chokki, M., Eguchi, H., Hamamura, I., Mitsuhashi, H. & Kamimura, T. Human airway trypsin-like protease induces amphiregulin release through a mechanism involving protease-activated receptor-2-mediated ERK activation and TNF alpha-converting enzyme activity in airway epithelial cells. FEBS J. 272, 6387–6399 (2005).
Li, Y. et al. Activation of protease-activated receptor-2 disrupts vaginal epithelial barrier function. Cell Biol. Int. 38, 1247–1251 (2014).
Papa, A. et al. Rad regulation of CaV1.2 channels controls cardiac fight-or-flight response. Nat. Cardiovasc Res 1, 1022–1038 (2022).
Antzelevitch, C. et al. Loss-of-function mutations in the cardiac calcium channel underlie a new clinical entity characterized by ST-segment elevation, short QT intervals, and sudden cardiac death. Circulation 115, 442–449 (2007).
Moras, E. et al. Genetic and molecular mechanisms in Brugada syndrome. Cells 12, 1791 (2023).
Watanabe, H. & Minamino, T. Genetics of Brugada syndrome. J. Hum. Genet. 61, 57–60 (2016).
Andrade, A. et al. Genetic associations between voltage-gated calcium channels and psychiatric disorders. Int. J. Mol. Sci. 20, 3537 (2019).
Flenady, V. et al. Calcium channel blockers for inhibiting preterm labour and birth. Cochrane Database Syst. Rev. 2014, CD002255 (2014).
Gáspár, R. & Hajagos-Tóth, J. Calcium channel blockers as tocolytics: principles of their actions, adverse effects and therapeutic combinations. Pharmaceuticals 6, 689–699 (2013).
Wray, S., Prendergast, C. & Arrowsmith, S. Calcium-activated chloride channels in myometrial and vascular smooth muscle. Front. Physiol. 12, 751008 (2021).
Conde-Agudelo, A., Romero, R. & Kusanovic, J. P. Nifedipine in the management of preterm labor: a systematic review and metaanalysis. Am. J. Obstet. Gynecol. 204, 134.e1–20 (2011).
Kuć, P., Laudański, P., Kowalczuk, O., Chyczewski, L. & Laudański, T. Expression of selected genes in preterm premature rupture of fetal membranes. Acta Obstet. Gynecol. Scand. 91, 936–943 (2012).
Trinchieri, G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. Immunol. 3, 133–146 (2003).
Tait Wojno, E. D., Hunter, C. A. & Stumhofer, J. S. The Immunobiology of the Interleukin-12 Family: Room for Discovery. Immunity 50, 851–870 (2019).
Gomez-Lopez, N. et al. The immunobiology of preterm labor and birth: intra-amniotic inflammation or breakdown of maternal-fetal homeostasis. Reproduction 164, R11–R45 (2022).
Hollier, L. M., Rivera, M. K., Henninger, E., Gilstrap, L. C. 3rd & Marshall, G. D. Jr. T helper cell cytokine profiles in preterm labor. Am. J. Reprod. Immunol. 52, 192–196 (2004).
Castle, P. E. et al. Cervical concentrations of interleukin-10 and interleukin-12 do not correlate with plasma levels. J. Clin. Immunol. 22, 23–27 (2002).
Gravitt, P. E. et al. Correlates of IL-10 and IL-12 concentrations in cervical secretions. J. Clin. Immunol. 23, 175–183 (2003).
Hildesheim, A. et al. Cytokine and immunoglobulin concentrations in cervical secretions: reproducibility of the Weck-cel collection instrument and correlates of immune measures. J. Immunol. Methods 225, 131–143 (1999).
Crowley-Nowick, P. A. et al. Cytokine profile in genital tract secretions from female adolescents: impact of human immunodeficiency virus, human papillomavirus, and other sexually transmitted pathogens. J. Infect. Dis. 181, 939–945 (2000).
Parrettini, S., Caroli, A. & Torlone, E. Nutrition and Metabolic Adaptations in Physiological and Complicated Pregnancy: Focus on Obesity and Gestational Diabetes. Front. Endocrinol. 11, 611929 (2020).
Jovandaric, M. Z., Babic, S., Raus, M. & Medjo, B. The importance of metabolic and environmental factors in the occurrence of oxidative stress during pregnancy. Int. J. Mol. Sci. 24, 11964 (2023).
Socha, M. W., Flis, W., Pietrus, M., Wartęga, M. & Stankiewicz, M. Signaling pathways regulating human cervical ripening in preterm and term delivery. Cells 11, 3690 (2022).
Yockey, L. J. & Iwasaki, A. Interferons and proinflammatory cytokines in pregnancy and fetal development. Immunity 49, 397–412 (2018).
Romero, R., Erez, O. & Espinoza, J. Intrauterine infection, preterm labor, and cytokines. J. Soc. Gynecol. Investig. 12, 463–465 (2005).
Yellon, S. M. Immunobiology of cervix ripening. Front. Immunol. 10, 3156 (2019).
Mendelson, C. R., Montalbano, A. P. & Gao, L. Fetal-to-maternal signaling in the timing of birth. J. Steroid Biochem. Mol. Biol. 170, 19–27 (2017).
Strauss, J. F. et al. Spontaneous preterm birth: advances toward the discovery of genetic predisposition. Am. J. Obstet. Gynecol. 218, 294–314.e2 (2018).
Romero, R., Dey, S. K. & Fisher, S. J. Preterm labor: One syndrome, many causes. Science 345, 760–765 (2014).
Myers, K. M. et al. The mechanical role of the cervix in pregnancy. J. Biomech. 48, 1511–1523 (2015).
Friedman, N. P., Banich, M. T. & Keller, M. C. Twin studies to GWAS: there and back again. Trends Cogn. Sci. 25, 855–869 (2021).
Plomin, R., DeFries, J. C., Knopik, V. S. & Neiderhiser, J. M. Behavioral Genetics. (Worth Publishers, 2012).
Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015).
Parry, L. J. & Vodstrcil, L. A. Relaxin physiology in the female reproductive tract during pregnancy. Adv. Exp. Med. Biol. 612, 34–48 (2007).
Goldsmith, L. T. & Weiss, G. Relaxin in human pregnancy. Ann. N. Y. Acad. Sci. 1160, 130–135 (2009).
Finlin, B. S. et al. RERG is a novel ras-related, estrogen-regulated and growth-inhibitory gene in breast cancer. J. Biol. Chem. 276, 42259–42267 (2001).
Key, M. D., Andres, D. A., Der, C. J. & Repasky, G. A. Characterization of RERG: an estrogen-regulated tumor suppressor gene. Methods Enzymol. 407, 513–527 (2006).
Deschênes, J., Bourdeau, V., White, J. H. & Mader, S. Regulation of GREB1 transcription by estrogen receptor alpha through a multipartite enhancer spread over 20 kb of upstream flanking sequences. J. Biol. Chem. 282, 17335–17339 (2007).
Mohammed, H. et al. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 3, 342–349 (2013).
Ozasa, H., Tominaga, T., Nishimura, T. & Takeda, T. Lysyl oxidase activity in the mouse uterine cervix is physiologically regulated by estrogen. Endocrinology 109, 618–621 (1981).
Akins, M. L., Luby-Phelps, K., Bank, R. A. & Mahendroo, M. Cervical softening during pregnancy: regulated changes in collagen cross-linking and composition of matricellular proteins in the mouse. Biol. Reprod. 84, 1053–1062 (2011).
Mahendroo, M. Cervical remodeling in term and preterm birth: insights from an animal model. Reproduction 143, 429–438 (2012).
Argraves, W. S., Tran, H., Burgess, W. H. & Dickerson, K. Fibulin is an extracellular matrix and plasma glycoprotein with repeated domain structure. J. Cell Biol. 111, 3155–3164 (1990).
Clinton, G. M. et al. Estrogens increase the expression of fibulin-1, an extracellular matrix protein secreted by human ovarian cancer cells. Proc. Natl. Acad. Sci. Usa. 93, 316–320 (1996).
Moll, F. et al. Estrogen induction and overexpression of fibulin-1C mRNA in ovarian cancer cells. Oncogene 21, 1097–1107 (2002).
Yip, C., Foidart, P., Noël, A. & Sounni, N. E. MT4-MMP: The GPI-anchored membrane-type matrix metalloprotease with multiple functions in diseases. Int. J. Mol. Sci. 20, 354 (2019).
de Vega, S., Iwamoto, T. & Yamada, Y. Fibulins: multiple roles in matrix structures and tissue functions. Cell. Mol. Life Sci. 66, 1890–1902 (2009).
Hao, Y. et al. Discovery and validation of FBLN1 and ANT3 as potential biomarkers for early detection of cervical cancer. Cancer Cell Int 21, 125 (2021).
Modi, B. P. et al. Rare mutations and potentially damaging missense variants in genes encoding fibrillar collagens and proteins involved in their production are candidates for risk for preterm premature rupture of membranes. PLoS One 12, e0174356 (2017).
Sizova, O., John, L. S., Ma, Q. & Molldrem, J. J. Multi-faceted role of LRP1 in the immune system. Front. Immunol. 14, 1166189 (2023).
Haas, J. et al. LRP1b shows restricted expression in human tissues and binds to several extracellular ligands, including fibrinogen and apoE-carrying lipoproteins. Atherosclerosis 216, 342–347 (2011).
Oxvig, C. The role of PAPP-A in the IGF system: Location, location, location. J. Cell Commun. Signal. 9, 177–187 (2015).
Kirkegaard, I., Uldbjerg, N. & Oxvig, C. Biology of pregnancy-associated plasma protein-A in relation to prenatal diagnostics: an overview. Acta Obstet. Gynecol. Scand. 89, 1118–1125 (2010).
Smith, G. C. S. et al. Early-pregnancy origins of low birth weight. Nature 417, 916 (2002).
Solovieff, N., Cotsapas, C., Lee, P. H., Purcell, S. M. & Smoller, J. W. Pleiotropy in complex traits: challenges and strategies. Nat. Rev. Genet. 14, 483–495 (2013).
Acknowledgements
This research was supported by the Perinatology Research Branch, Division of Obstetrics and Maternal-Fetal Medicine, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, United States Department of Health and Human Services (NICHD/NIH/DHHS) (Contract No. HHSN275201300006C). R.R. has contributed to this work as part of his official duties as an employee of the United States Federal Government. A.L.T., N.G.L., and S.S.H. were also supported by the Wayne State University Perinatal Initiative in Maternal, Perinatal and Child Health. Additional support was provided from the March of Dimes Prematurity Research Center at the University of Pennsylvania (22-FY18-812).
Author information
Authors and Affiliations
Contributions
T.P.Y., J.F.S, and R.R. conceived and designed the study. H.M.W, T.P.Y., A.L.T., S.J.L., and B.T.W. contributed to analysis of the data. R.R., S.S.H., T.C., S.B., N.G.L., and P.C. acquired the data and biospecimens. T.P.Y. and H.M.W. developed the manuscript draft. J.F.S, B.T.W., A.L.T., N.G.L., and R.R. critically revised the article. All authors gave final approval of the version to be published.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Medicine thanks Uri Amikam and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
*The Perinatology Research Branch, NICHD/NIH/DHHS, has been renamed as the Pregnancy Research Branch, NICHD/NIH/DHHS.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wolf, H.M., Webb, B.T., Strauss, J.F. et al. The genetic architecture of cervical length is shared with spontaneous preterm birth risk. Commun Med 5, 352 (2025). https://doi.org/10.1038/s43856-025-01078-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43856-025-01078-0