Abstract
Background
Postoperative pancreatic fistula (POPF) is the major driver of postoperative morbidity after pancreatoduodenectomy (PD) and a healthcare issue. In patients with pancreatic tumors the occurrence of POPF could lead to a complete failure of the oncologic strategy by delaying or annihilating the delivery of the indicated adjuvant chemotherapy. However, current preoperative prediction models lack precision. This study aimed to determine the ability of a high dimensional analysis of the patient’s peripheral immune system before PD to predict POPF.
Methods
Twenty-two patients in the prospective IMMUNOPANC trial (NCT03978702) underwent PD. Blood samples collected preoperatively were analyzed by combining single-cell mass cytometry and a sparse machine-learning pipeline, Stabl, to identify the most relevant POPF-predictive features. The logistic regression model output was evaluated using a five-fold cross-validation procedure.
Results
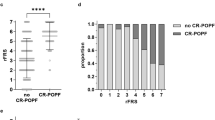
Eight (36%) patients experience POPF (grade B, n = 7; grade C, n = 1). The multivariable predictive model includes 11 features—six natural killer, three CD8+ T, and two CD4+ T lymphocyte cell clusters—revealing a preoperative POPF lymphocyte signature (Pancreatic Fistula Lymphocyte Signature, PFLS). The Stabl algorithm identifies a predictive model classifying POPF patients with high performance (area under the receiver operating characteristic curve=0.81, P = 2.04e-02).
Conclusions
In summary, preoperative circulating immune-cell composition can predict POPF in patients undergoing PD. The clinical application of the PFLS may enable the early identification of patients at high risk before pancreatic surgery, giving clinicians the opportunity to anticipate and mitigate POPF risk through tailored strategies in pre-, intra-, and post-operative settings.

Plain language summary
Pancreatic surgery is the cornerstone of treatment for patients with pancreatic tumors, but a serious complication called postoperative pancreatic fistula (POPF) often occurs. POPF can worsen recovery and delay critical chemotherapy, sometimes causing cancer treatment to fail. Current tools to predict which patients are at risk for POPF are not accurate enough. In this study, we tested whether analyzing immune cells in the blood with machine learning could better predict POPF before surgery. We found that this approach accurately identified patients at high risk for POPF. Early recognition of patients at risk of POPF could allow physicians to anticipate and reduce said risk through tailored strategies before, during, and after surgery, ultimately improving recovery and access to timely cancer treatment.
Similar content being viewed by others
Data availability
The datasets generated and/or analyzed during the current study are not publicly available because of patient privacy concerns but are available from the corresponding author upon reasonable request. The timeframe for response is within two weeks. Use of data is controlled by data transfer agreement. The source data for the figures is accessible in the corresponding supplementary data. Supplementary Data 1 was generated with the non-supervised dimensionality reduction algorithm of h-SNE implemented in Cytosplore (V2.2.1)29, and represents the Fig. 2. Of note, the workflow from PBMCs to h-SNE clustering has been previously reported22. The unit is the proportion of cells count. Supplementary Data 2, 3 and 4 were used to generate Fig. 3A. Supplementary Data 2 contains all immune clusters and the POPF outcome. Blood samples collected the day before surgery were used to calculate the absolute cell count for each immune cluster (unit: 10⁶ cells/L). Supplementary Data 3 contains the multiomic Stabl+logistic regression cross-validation for all the features, and Supplementary Data 4 contains the multiomic Stabl+logistic regression validation coefficients for the 11 clusters of the signature. Supplementary Data 5 was used to generate Fig. 3B, C, as well as Fig. 4. It contains the POPF outcome, the ua-FRS index, the PFLS result, the clinical and biological values for each patient.
Code availability
The analyses, results, and figures reported in this study were generated using Biomics Software, a proprietary machine-learning platform developed and maintained by SurgeCare. Consequently, the exact implementation code used in this study is not publicly available, as it forms part of a commercial software product. Importantly, Biomics Software is built upon established, openly available machine-learning methodologies. In particular, the core feature-selection methodology used in this study relies on the Stabl algorithm, which is fully described in the literature and whose reference implementation is publicly available (Hédou, J., Marić, I., Bellan, G. et al. Discovery of sparse, reliable omic biomarkers with Stabl. Nat Biotechnol 42, 1581–1593 (2024). https://doi.org/10.1038/s41587-023-02033-x). Additional foundational components rely on widely used open-source machine-learning libraries (e.g., Python-based scientific and machine-learning frameworks), which are freely accessible online. The full analytical workflow, including model design, training procedures, validation strategy, and evaluation metrics, is described in detail in the Methods section to ensure methodological transparency and reproducibility. Due to the proprietary nature of the Biomics Software platform, we are unable to deposit the full application code in a public repository or assign a DOI. However, access to the software for editorial or reviewer verification can be provided upon reasonable request, subject to appropriate confidentiality and usage agreements.
References
Ma, L. W. et al. The cost of postoperative pancreatic fistula versus the cost of pasireotide: results from a prospective randomized trial. Ann. Surg. 265, 11–16 (2017).
Williamsson, C., Ansari, D., Andersson, R. & Tingstedt, B. Postoperative pancreatic fistula-impact on outcome, hospital cost and effects of centralization. HPB (Oxford) 19, 436–442 (2017).
Wellner, U. F. et al. A simple scoring system based on clinical factors related to pancreatic texture predicts postoperative pancreatic fistula preoperatively. HPB (Oxford) 12, 696–702 (2010).
Yamamoto, Y. et al. A preoperative predictive scoring system for postoperative pancreatic fistula after pancreaticoduodenectomy. World J. Surg. 35, 2747–2755 (2011).
Roberts, K. J. et al. Scoring system to predict pancreatic fistula after pancreaticoduodenectomy: a UK multicenter study. Ann. Surg. 261, 1191–1197 (2015).
Nishida, Y. et al. Preoperative sarcopenia strongly influences the risk of postoperative pancreatic fistula formation after pancreaticoduodenectomy. J. Gastrointest. Surg. 20, 1586–1594 (2016).
Schuh, F. et al. A Simple Classification of Pancreatic Duct Size and Texture Predicts postoperative Pancreatic Fistula: a classification of the International Study Group of Pancreatic Surgery (ISGPS). Ann. Surg. 277, e597–e608 (2023).
Callery, M. P., Pratt, W. B., Kent, T. S., Chaikof, E. L. & Vollmer, C. M. A prospectively validated clinical risk score accurately predicts pancreatic fistula after pancreatoduodenectomy. J. Am. Coll. Surg. 216, 1–14 (2013).
Nimptsch, U., Krautz, C., Weber, G. F., Mansky, T. & Grützmann, R. Nationwide in-hospital mortality following pancreatic surgery in Germany is higher than anticipated. Ann. Surg. 264, 1082–1090 (2016).
Farges, O. et al. The theory and practice of pancreatic surgery in France. Ann. Surg. 266, 797–804 (2017).
El Amrani, M. et al. Referring patients to expert centers after pancreatectomy is too late to improve outcome. Inter-hospital transfer analysis in nationwide study of 19,938 patients. Ann. Surg. 272, 723–730 (2020).
Mungroop, T. H. et al. Updated alternative fistula risk score (ua-FRS) to include minimally invasive pancreatoduodenectomy: pan-European validation. Ann. Surg. 273, 334–340 (2021).
Casciani, F., Bassi, C. & Vollmer, C. M. Decision points in pancreatoduodenectomy: insights from the contemporary experts on prevention, mitigation, and management of postoperative pancreatic fistula. Surgery 170, 889–909 (2021).
Fong, Z. V. et al. Early drain removal—the middle ground between the Drain versus No Drain debate in patients undergoing pancreaticoduodenectomy: a prospective validation study. Ann. Surg. 262, 378–383 (2015).
Van Hilst, J. et al. The inflammatory response after laparoscopic and open pancreatoduodenectomy and the association with complications in a multicenter randomized controlled trial. HPB 21, 1453–1461 (2019).
Garnier, J. et al. Establishment and external validation of neutrophil-to-lymphocyte ratio in excluding postoperative pancreatic fistula after pancreatoduodenectomy. BJS Open 7, zrac124 (2023).
Søreide, K., Healey, A. J., Mole, D. J. & Parks, R. W. Pre-, peri- and post-operative factors for the development of pancreatic fistula after pancreatic surgery. HPB 21, 1621–1631 (2019).
Verdonk, F. et al. Measuring the human immune response to surgery: multiomics for the prediction of postoperative outcomes. Curr. Opin. Crit. Care 27, 717–725 (2021).
Rumer, K. K. et al. Integrated single-cell and plasma proteomic modeling to predict surgical site complications: a prospective cohort study. Ann. Surg. 275, 582–590 (2022).
Fragiadakis, G. K. et al. Patient-specific immune states before surgery are strong correlates of surgical recovery. Anesthesiology 123, 1241–1255 (2015).
Verdonk, F. et al. An immune signature of postoperative cognitive decline: a prospective cohort study.Int. J. Surg. 110, 7749–7762 (2024).
Garnier, J. et al. Immediate variations in high-dimensional circulating immune cells following pancreatectomy. Clin. Transl. Immunol. 14, e70059 (2025).
Collins, G. S., Reitsma, J. B., Altman, D. G. & Moons, K. G. M. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD Statement. BMC Med. 13, 1 https://doi.org/10.1186/s12916-014-0241-z (2015).
Bassi, C. et al. The 2016 update of the International Study Group (ISGPS) definition and grading of postoperative pancreatic fistula: 11 years after. Surgery 161, 584–591 (2017).
Halloran, C. M. et al. A multicenter, randomized, double-blinded, clinical trial comparing Cattell-Warren and Blumgart anastomoses following partial pancreatoduodenectomy: PANasta trial. Ann. Surg. Open 3, e198 (2022).
Turrini, O. & Delpero, J. R. Omental flap for vessel coverage during pancreaticoduodenectomy. A Modified Technique. J. Chir. (Paris) 146, 545–548 (2009).
Pessaux, P. et al. External pancreatic duct stent decreases pancreatic fistula rate after pancreaticoduodenectomy: prospective multicenter randomized trial. Ann. Surg. 253, 879–885 (2011).
Ecker, B. L. et al. Characterization and optimal management of high-risk pancreatic anastomoses during pancreatoduodenectomy. Ann. Surg. 267, 608–616 (2018).
Van Unen, V. et al. Visual analysis of mass cytometry data by hierarchical stochastic neighbour embedding reveals rare cell types. Nat. Commun. 8, 1740 (2017).
Hédou, J. et al. Discovery of sparse, reliable omic biomarkers with Stabl. Nat. Biotechnol. 42, 1581–1593 (2024).
Ahmed, R. et al. CD57+ Memory T cells proliferate in vivo. Cell Rep 33, 108501 (2020).
Björkström, N. K. et al. Elevated numbers of FcγRIIIA+ (CD16+) effector CD8 T cells with NK cell-like function in chronic hepatitis C virus infection. J. Immunol. 181, 4219–4228 (2008).
Freuchet, A. et al. Identification of human exTreg cells as CD16+CD56+ cytotoxic CD4+ T cells. Nat. Immunol. 24, 1748–1761 (2023).
Hirono, S. et al. Risk factors for pancreatic fistula grade C after pancreatoduodenectomy: a large prospective, multicenter Japan-Taiwan collaboration study. J. Hepatobiliary Pancreat Sci. 27, 622–631 (2020).
Bassi, C. et al. Pancreatoduodenectomy at the Verona Pancreas Institute: the evolution of indications, surgical techniques, and outcomes: a retrospective analysis of 3000 consecutive cases. Ann. Surg. 276, 1029–1038 (2022).
McMillan, M. T. et al. The characterization and prediction of ISGPF Grade C fistulas following pancreatoduodenectomy. J. Gastrointest. Surg. 20, 262–276 (2016).
Hank, T. et al. Association between pancreatic fistula and long-term survival in the era of neoadjuvant chemotherapy. JAMA Surg. 154, 943 (2019).
Reinke, S. et al. Terminally differentiated CD8+ T cells negatively affect bone regeneration in humans. Sci. Transl. Med. 5, 177ra36 (2013).
Slade, M., Simmons, R., Yunis, E. & Greenberg, L. Immunodepression after major surgery in normal patients. Surgery 8, 363–372 (1975).
Cohen, J. T., Charpentier, K. P., Miner, T. J., Cioffi, W. G. & Beard, R. E. Lymphopenia following pancreaticoduodenectomy is associated with pancreatic fistula formation. Ann. Hepatobiliary Pancreat Surg. 25, 242–250 (2021).
Chretien, A. S. et al. Natural killer defective maturation is associated with adverse clinical outcome in patients with acute myeloid leukemia. Front. Immunol. 8, 573 (2017).
Conway Morris, A. et al. Cell-surface signatures of immune dysfunction risk-stratify critically ill patients: INFECT study. Intensive Care Med. 44, 627–635 (2018).
Sharma, P. & Allison, J. P. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell 161, 205–214 (2015).
Werba, G. et al. Single-cell RNA sequencing reveals the effects of chemotherapy on human pancreatic adenocarcinoma and its tumor microenvironment. Nat. Commun. 14, 797 (2023).
Middelburg, J. et al. The MHC-E peptide ligands for checkpoint CD94/NKG2A are governed by inflammatory signals, whereas LILRB1/2 receptors are peptide indifferent. Cell Rep. 42, 113516 (2023).
Hyung, J. et al. Tumor immune-gene expression profiles and peripheral immune phenotypes associated with clinical outcomes of locally advanced pancreatic cancer following FOLFIRINOX. ESMO Open 7, 100484 (2022).
Shinde, R. S. et al. External validation and comparison of the original, alternative and updated-alternative fistula risk scores for the prediction of postoperative pancreatic fistula after pancreatoduodenectomy. Pancreatology 20, 751–756 (2020).
Gu, Z. et al. Development and validation of a novel nomogram to predict postoperative pancreatic fistula after pancreatoduodenectomy using lasso-logistic regression: an international multi-institutional observational study. Int. J. Surg. 109, 4027–4040 (2023).
Ashraf Ganjouei, A. et al. A machine learning approach to predict postoperative pancreatic fistula after pancreaticoduodenectomy using only preoperatively known data. Ann. Surg. Oncol. 30, 7738–7747 (2023).
Verma, A. et al. Machine learning-based prediction of postoperative pancreatic fistula following pancreaticoduodenectomy. Ann. Surg. 280, 325–331 (2024).
Ingwersen, E. W. et al. Radiomics preoperative-Fistula risk score (RAD-FRS) for pancreatoduodenectomy: development and external validation. BJS Open 7, zrad100 (2023).
Mišić, V. V., Gabel, E., Hofer, I., Rajaram, K. & Mahajan, A. Machine learning prediction of postoperative emergency department hospital readmission. Anesthesiology 132, 968–980 (2020).
Durand, X. et al. Predicthor: AI-powered predictive risk model for 30-day mortality and 30-day complications in patients undergoing thoracic surgery for lung cancer. Ann. Surg. Open 6, e578 (2025).
Tarvainen, T., Sirén, J., Kokkola, A. & Sallinen, V. Effect of hydrocortisone vs pasireotide on pancreatic surgery complications in patients with high risk of pancreatic fistula: a randomized clinical trial. JAMA Surg 155, 291–298 (2020).
Palen, A., Garnier, J., Delpero, J. R., Turrini, O. & Ewald, J. Protective peritoneal patch for arteries during pancreatoduodenectomy: good value for money. Langenbecks Arch Surg 407, 377–382 (2022).
Smits, F. J. et al. Algorithm-based care versus usual care for the early recognition and management of complications after pancreatic resection in the Netherlands: an open-label, nationwide, stepped-wedge cluster-randomised trial. Lancet 399, 1867–1875 (2022).
De Luca, R. et al. Immunonutrition and prehabilitation in pancreatic cancer surgery: A new concept in the era of ERAS® and neoadjuvant treatment. Eur. J. Surg. Oncol. 49, 542–549 (2023).
Barberan-Garcia, A. et al. Personalised prehabilitation in high-risk patients undergoing elective major abdominal surgery: a randomized blinded controlled trial. Ann. Surg. 267, 50–56 (2018).
Cambriel, A. et al. Immune modulation by personalized vs standard prehabilitation before major surgery: a randomized clinical trial. JAMA Surg. 161, 20–30 (2026).
Balzano, G. et al. Total pancreatectomy with islet autotransplantation as an alternative to high-risk pancreatojejunostomy after pancreaticoduodenectomy: a prospective randomized trial. Ann. Surg. 277, 894–903 (2023).
Stoop, T. F. et al. Systematic review and meta-analysis of the role of total pancreatectomy as an alternative to pancreatoduodenectomy in patients at high risk for postoperative pancreatic Fistula: is it a justifiable indication? Ann Surg. 278, e702–e711 (2023).
Davern, M. et al. PD-1 blockade attenuates surgery-mediated immunosuppression and boosts Th1 immunity perioperatively in oesophagogastric junctional adenocarcinoma. Front. Immunol. 14, 1150754 (2023).
Hotchkiss, R. S. et al. Immune checkpoint inhibition in sepsis: a Phase 1b randomized, placebo-controlled, single ascending dose study of antiprogrammed cell death-ligand 1 antibody (BMS-936559). Crit. Care Med. 47, 632–642 (2019).
Becht, E. et al. Reverse-engineering flow-cytometry gating strategies for phenotypic labelling and high-performance cell sorting. Bioinformatics 35, 301–308 (2019).
De Graaf, N. et al. Minimally invasive versus open pancreatoduodenectomy for resectable neoplasms. NEJM Evid. 4, EVIDoa2500045 (2025).
Maslove, D. M. et al. Redefining critical illness. Nat. Med. 28, 1141–1148 (2022).
Acknowledgements
This work was supported by the Fondation ARC (grant ARC#2022-00154 for ASC), the Groupement d’intérêt scientifique -Infrastructures pour la Biologie, la Santé et l’Agronomie (GIS IBiSA) and the National Institute of Health (grant R35GM137936 for BG). The team “Immunity and Cancer” was labeled “Equipe Fondation pour la Recherche Médicale (FRM) #2018-00198” (for D.O.).
Author information
Authors and Affiliations
Contributions
Conceptualization: Jonathan Garnier, Olivier Turrini, Anne-Sophie Chrétien and Daniel Olive; Data curation: Jonathan Garnier, Olivier Turrini, Marie Sarah Rouvière, and Amira Ben-Amara; Formal analysis: Jonathan Garnier, Anne-Sophie Chrétien, Marie Sarah Rouvière, Amira Ben-Amara, Gregoire Bellan, and Xavier Durand; Investigation: Jonathan Garnier, Olivier Turrini, and Anne-Sophie Chrétien; Methodology: Jonathan Garnier, Olivier Turrini, Anne-Sophie Chrétien, Daniel Olive, Gregoire Bellan, and Xavier Durand; Project administration: Jonathan Garnier, Olivier Turrini, and Caroline Gouarné; Resources: Jonathan Garnier, Olivier Turrini, Anaïs Palen, Jacques Ewald, Caroline Gouarné, Anne-Sophie Chrétien, Daniel Olive, Franck Verdonk, and Brice Gaudilliere; Software: Jonathan Garnier, Anne-Sophie Chrétien, Benjamin Choisy, Gregoire Bellan, and Xavier Durand; Writing—original draft: Jonathan Garnier, Anne-Sophie Chrétien, Gregoire Bellan, and Xavier Durand; Writing—review & editing: all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Medicine thanks Vera Hartman and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Garnier, J., Bellan, G., Palen, A. et al. Preoperative lymphocyte signature predicts pancreatic fistula after pancreatoduodenectomy. Commun Med (2026). https://doi.org/10.1038/s43856-026-01422-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43856-026-01422-y