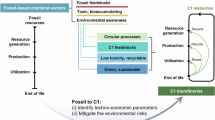
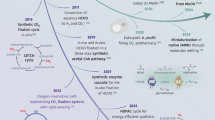
Abstract
Life—ranging from cellular processes to the complexities of modern societies—requires a diverse array of chemicals to function. Whereas humans have become adept at synthesizing incredible chemical diversity over the past two centuries, these practices still rely on the use (and breakdown) of fossil resources. However, the challenge of climate change makes it clear that sustainable chemical synthesis requires alternative methods and substrates. The growing abundance of carbonaceous gases in the atmosphere (in particular, carbon dioxide and methane) could serve as feedstocks for such a sustainable synthesis transition, and biological systems are adept at converting one-carbon (C1) compounds into more complex molecules. This Review discusses recent developments and future opportunities for the biosynthesis of chemicals from C1 substrates via cellular and cell-free systems. In addition to the diverse range of products synthesized using natural or designed C1 conversion pathways in vivo or in vitro, we discuss the benefits of spatio-temporal organization and hybrid catalysis to increase the efficiency of enzymatic chemical synthesis from C1 substrates.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Liu, Z., Wang, K., Chen, Y., Tan, T. & Nielsen, J. Third-generation biorefineries as the means to produce fuels and chemicals from CO2. Nat. Catal. 3, 274–288 (2020).
LeClerc, H. O. et al. The CO2 tree: the potential for carbon dioxide utilization pathways. ACS Sustain. Chem. Eng. 13, 5–29 (2025).
Vlaeminck, E. et al. Pressure fermentation to boost CO2-based poly(3-hydroxybutyrate) production using Cupriavidus necator. Bioresour. Technol. 408, 131162 (2024).
Liew, F. et al. Gas fermentation—a flexible platform for commercial scale production of low-carbon-fuels and chemicals from waste and renewable feedstocks. Front. Microbiol. 7, 694 (2016).
Santos-Merino, M., Singh, A. K. & Ducat, D. C. New applications of synthetic biology tools for cyanobacterial metabolic engineering. Front. Bioeng. Biotechnol. 7, 33 (2019).
Chen, F. Y.-H., Jung, H.-W., Tsuei, C.-Y. & Liao, J. C. Converting Escherichia coli to a synthetic methylotroph growing solely on methanol. Cell 182, 933–946.e14 (2020).
Gleizer, S. et al. Conversion of Escherichia coli to generate all biomass carbon from CO2. Cell 179, 1255–1263.e12 (2019).
Keller, P. et al. Generation of an Escherichia coli strain growing on methanol via the ribulose monophosphate cycle. Nat. Commun. 13, 5243 (2022).
Liew, F. E. et al. Carbon-negative production of acetone and isopropanol by gas fermentation at industrial pilot scale. Nat. Biotechnol. 40, 335–344 (2022).
Nangle, S. N. et al. Valorization of CO2 through lithoautotrophic production of sustainable chemicals in Cupriavidus necator. Metab. Eng. 62, 207–220 (2020).
Schwander, T., Schada von Borzyskowski, L., Burgener, S., Cortina, N. S. & Erb, T. J. A synthetic pathway for the fixation of carbon dioxide in vitro. Science 354, 900–904 (2016).
Bogorad, I. W., Lin, T.-S. & Liao, J. C. Synthetic non-oxidative glycolysis enables complete carbon conservation. Nature 502, 693–697 (2013).
Liu, C., Colón, B. C., Ziesack, M., Silver, P. A. & Nocera, D. G. Water splitting–biosynthetic system with CO2 reduction efficiencies exceeding photosynthesis. Science 352, 1210–1213 (2016).
Diehl, C., Gerlinger, P. D., Paczia, N. & Erb, T. J. Synthetic anaplerotic modules for the direct synthesis of complex molecules from CO2. Nat. Chem. Biol. 19, 168–175 (2023).
Cai, T. et al. Cell-free chemoenzymatic starch synthesis from carbon dioxide. Science 373, 1523–1527 (2021).
O’Keeffe, S. et al. Bringing carbon to life via one-carbon metabolism. Trends Biotechnol. 43, 572–585 (2025).
Hudson, E. P. The Calvin Benson cycle in bacteria: new insights from systems biology. Semin. Cell Dev. Biol. 155, 71–83 (2024).
Bar-Even, A., Noor, E. & Milo, R. A survey of carbon fixation pathways through a quantitative lens. J. Exp. Bot. 63, 2325–2342 (2012).
Sulis, D. B. et al. Multiplex CRISPR editing of wood for sustainable fiber production. Science 381, 216–221 (2023).
Santos-Merino, M., Yun, L. & Ducat, D. C. Cyanobacteria as cell factories for the photosynthetic production of sucrose. Front. Microbiol. 14, 1126032 (2023).
Sanford, P. A. & Woolston, B. M. Expanding the genetic engineering toolbox for the metabolically flexible acetogen Eubacterium limosum. J. Ind. Microbiol. Biotechnol. 49, kuac019 (2022).
Pham, D. N., Nguyen, A. D. & Lee, E. Y. Outlook on engineering methylotrophs for one-carbon-based industrial biotechnology. Chem. Eng. J. 449, 137769 (2022).
Bysani, V. R., Alam, A. S., Bar-Even, A. & Machens, F. Engineering and evolution of the complete reductive glycine pathway in Saccharomyces cerevisiae for formate and CO2 assimilation. Metab. Eng. 81, 167–181 (2024).
Kim, S. et al. Growth of E. coli on formate and methanol via the reductive glycine pathway. Nat. Chem. Biol. 16, 538–545 (2020).
Guo, Y. et al. Engineering yeasts to co-utilize methanol or formate coupled with CO2 fixation. Metab. Eng. 84, 1–12 (2024).
Gassler, T. et al. The industrial yeast Pichia pastoris is converted from a heterotroph into an autotroph capable of growth on CO2. Nat. Biotechnol. 38, 210–216 (2020).
Ben Nissan, R. et al. Autotrophic growth of Escherichia coli is achieved by a small number of genetic changes. eLife 12, RP88793 (2024).
Gassler, T., Baumschabl, M., Sallaberger, J., Egermeier, M. & Mattanovich, D. Adaptive laboratory evolution and reverse engineering enhances autotrophic growth in Pichia pastoris. Metab. Eng. 69, 112–121 (2022).
Reiter, M. A. et al. A synthetic methylotrophic Escherichia coli as a chassis for bioproduction from methanol. Nat. Catal. 7, 560–573 (2024).
Jiang, W. et al. Metabolic engineering strategies to enable microbial utilization of C1 feedstocks. Nat. Chem. Biol. 17, 845–855 (2021).
Schulz-Mirbach, H., Dronsella, B., He, H. & Erb, T. J. Creating new-to-nature carbon fixation: a guide. Metab. Eng. 82, 12–28 (2024).
Erb, T. J., Jones, P. R. & Bar-Even, A. Synthetic metabolism: metabolic engineering meets enzyme design. Curr. Opin. Chem. Biol. 37, 56–62 (2017).
Bar-Even, A., Noor, E., Lewis, N. E. & Milo, R. Design and analysis of synthetic carbon fixation pathways. Proc. Natl Acad. Sci. USA 107, 8889–8894 (2010).
Pandit, A. V., Srinivasan, S. & Mahadevan, R. Redesigning metabolism based on orthogonality principles. Nat. Commun. 8, 15188 (2017).
Pandi, A. et al. A versatile active learning workflow for optimization of genetic and metabolic networks. Nat. Commun. 13, 3876 (2022).
Luo, S. et al. Construction and modular implementation of the THETA cycle for synthetic CO2 fixation. Nat. Catal. 6, 1228–1240 (2023).
Wu, C. et al. Acetyl-CoA synthesis through a bicyclic carbon-fixing pathway in gas-fermenting bacteria. Nat. Synth. 1, 615–625 (2022).
Chou, A., Lee, S. H., Zhu, F., Clomburg, J. M. & Gonzalez, R. An orthogonal metabolic framework for one-carbon utilization. Nat. Metab. 3, 1385–1399 (2021).
Dookeran, Z. A. & Nielsen, D. R. Systematic engineering of Synechococcus elongatus UTEX 2973 for photosynthetic production of L-lysine, cadaverine, and glutarate. ACS Synth. Biol. 10, 3561–3575 (2021).
Mishra, S., Perkovich, P. M., Mitchell, W. P., Venkataraman, M. & Pfleger, B. F. Expanding the synthetic biology toolbox of Cupriavidus necator for establishing fatty acid production. J. Ind. Microbiol. Biotechnol. 51, kuae008 (2024).
Xie, H., Kjellström, J. & Lindblad, P. Sustainable production of photosynthetic isobutanol and 3-methyl-1-butanol in the cyanobacterium Synechocystis sp. PCC 6803. Biotechnol. Biofuels Bioprod. 16, 134 (2023).
Yunus, I. S. et al. Improved bioproduction of 1-octanol using engineered Synechocystis sp. PCC 6803. ACS Synth. Biol. 10, 1417–1428 (2021).
Vögeli, B. et al. Cell-free prototyping enables implementation of optimized reverse β-oxidation pathways in heterotrophic and autotrophic bacteria. Nat. Commun. 13, 3058 (2022).
Ma, Z.-x. et al. Metabolomic analysis improves bioconversion of methanol to isobutanol in Methylorubrum extorquens AM1. Biotechnol. J. 16, 2000413 (2021).
Sonntag, F. et al. Engineering Methylobacterium extorquens for de novo synthesis of the sesquiterpenoid α-humulene from methanol. Metab. Eng. 32, 82–94 (2015).
Quynh Le, H. T., Anh Mai, D. H., Na, J.-G. & Lee, E. Y. Development of Methylorubrum extorquens AM1 as a promising platform strain for enhanced violacein production from co-utilization of methanol and acetate. Metab. Eng. 72, 150–160 (2022).
Golubova, D., Tansley, C., Su, H. & Patron, N. J. Engineering Nicotiana benthamiana as a platform for natural product biosynthesis. Curr. Opin. Plant Biol. 81, 102611 (2024).
Einhaus, A., Baier, T. & Kruse, O. Molecular design of microalgae as sustainable cell factories. Trends Biotechnol. 42, 728–738 (2024).
Einhaus, A. et al. Engineering a powerful green cell factory for robust photoautotrophic diterpenoid production. Metab. Eng. 73, 82–90 (2022).
Calgaro-Kozina, A. et al. Engineering plant synthetic pathways for the biosynthesis of novel antifungals. ACS Cent. Sci. 6, 1394–1400 (2020).
Barnum, C. R. et al. Engineered plants provide a photosynthetic platform for the production of diverse human milk oligosaccharides. Nat. Food 5, 480–490 (2024).
Zhang, J. et al. A microbial supply chain for production of the anti-cancer drug vinblastine. Nature 609, 341–347 (2022).
Messiha, H. L., Scrutton, N. S. & Leys, D. High-titer bio-styrene production afforded by whole-cell cascade biotransformation. ChemCatChem 15, e202201102 (2023).
Pyne, M. E. et al. A yeast platform for high-level synthesis of tetrahydroisoquinoline alkaloids. Nat. Commun. 11, 3337 (2020).
Sundaram, S. et al. A modular in vitro platform for the production of terpenes and polyketides from CO2. Angew. Chem. Int. Ed. 60, 16420–16425 (2021).
Flamholz, A. & Shih, P. M. Cell biology of photosynthesis over geologic time. Curr. Biol. 30, R490–R494 (2020).
Abrahamson, C. H., Palmero, B. J., Kennedy, N. W. & Tullman-Ercek, D. Theoretical and practical aspects of multienzyme organization and encapsulation. Annu. Rev. Biophys. 52, 553–572 (2023).
Dueber, J. E. et al. Synthetic protein scaffolds provide modular control over metabolic flux. Nat. Biotechnol. 27, 753–759 (2009).
Fernandez-Lorente, G. et al. Interfacially activated lipases against hydrophobic supports: effect of the support nature on the biocatalytic properties. Process Biochem. 43, 1061–1067 (2008).
Sánchez-Morán, H., Kaar, J. L. & Schwartz, D. K. Supra-biological performance of immobilized enzymes enabled by chaperone-like specific non-covalent interactions. Nat. Commun. 15, 2299 (2024).
Oskoei, V., Mathesh, M. & Yang, W. Enhancing substrate channeling with multi-enzyme architectures in hydrogen-bonded organic frameworks. Chem. Eur. J. 30, e202401256 (2024).
Gao, Y., Roberts, C. C., Toop, A., Chang, C. A. & Wheeldon, I. Mechanisms of enhanced catalysis in enzyme–DNA nanostructures revealed through molecular simulations and experimental analysis. ChemBioChem 17, 1430–1436 (2016).
Satagopan, S., Sun, Y., Parquette, J. R. & Tabita, F. R. Synthetic CO2-fixation enzyme cascades immobilized on self-assembled nanostructures that enhance CO2/O2 selectivity of RubisCO. Biotechnol. Biofuels 10, 175 (2017).
Fan, G. et al. Highly efficient carbon dioxide electroreduction via DNA-directed catalyst immobilization. JACS Au 4, 1413–1421 (2024).
Zhang, Y. et al. Protein–protein interactions and metabolite channelling in the plant tricarboxylic acid cycle. Nat. Commun. 8, 15212 (2017).
Wu, F. & Minteer, S. Krebs cycle metabolon: structural evidence of substrate channeling revealed by cross-linking and mass spectrometry. Angew. Chem. Int. Ed. 54, 1851–1854 (2015).
Steffens, L. et al. High CO2 levels drive the TCA cycle backwards towards autotrophy. Nature 592, 784–788 (2021).
Eun, C., Kekenes-Huskey, P. M., Metzger, V. T. & McCammon, J. A. A model study of sequential enzyme reactions and electrostatic channeling. J. Chem. Phys. 140, 105101 (2014).
Kummer, M. J. et al. Substrate channeling by a rationally designed fusion protein in a biocatalytic cascade. JACS Au 1, 1187–1197 (2021).
Yu, S. et al. Light-driven enzymatic nanosystem for highly selective production of formic acid from CO2. Chem. Eng. J. 420, 127649 (2021).
Breger, J. C. et al. Self assembling nanoparticle enzyme clusters provide access to substrate channeling in multienzymatic cascades. Nat. Commun. 14, 1757 (2023).
Kerfeld, C. A., Aussignargues, C., Zarzycki, J., Cai, F. & Sutter, M. Bacterial microcompartments. Nat. Rev. Microbiol. 16, 277–290 (2018).
Rae, B. D., Long, B. M., Badger, M. R. & Price, G. D. Functions, compositions, and evolution of the two types of carboxysomes: polyhedral microcompartments that facilitate CO2 fixation in cyanobacteria and some proteobacteria. Microbiol. Mol. Biol. Rev. 77, 357–379 (2013).
Dou, Z. et al. CO2 fixation kinetics of Halothiobacillus neapolitanus mutant carboxysomes lacking carbonic anhydrase suggest the shell acts as a diffusional barrier for CO2. J. Biol. Chem. 283, 10377–10384 (2008).
Flamholz, A. I. et al. Functional reconstitution of a bacterial CO2 concentrating mechanism in Escherichia coli. eLife 9, e59882 (2020).
Slininger Lee, M. F., Jakobson, C. M. & Tullman-Ercek, D. Evidence for improved encapsulated pathway behavior in a bacterial microcompartment through shell protein engineering. ACS Synth. Biol. 6, 1880–1891 (2017).
Lawrence, A. D. et al. Solution structure of a bacterial microcompartment targeting peptide and its application in the construction of an ethanol bioreactor. ACS Synth. Biol. 3, 454–465 (2014).
Kirst, H. et al. Toward a glycyl radical enzyme containing synthetic bacterial microcompartment to produce pyruvate from formate and acetate. Proc. Natl Acad. Sci. USA 119, e2116871119 (2022).
Küffner, A. M. et al. Acceleration of an enzymatic reaction in liquid phase separated compartments based on intrinsically disordered protein domains. ChemSystemsChem 2, e2000001 (2020).
Freeman Rosenzweig, E. S. et al. The eukaryotic CO2-concentrating organelle is liquid-like and exhibits dynamic reorganization. Cell 171, 148–162.e19 (2017).
Küffner, A. M. et al. Bottom-up reconstruction of minimal pyrenoids provides insights into the evolution and mechanisms of carbon concentration by EPYC1 proteins. Preprint at bioRxiv https://doi.org/10.1101/2024.06.28.601168 (2024).
Blikstad, C. et al. Identification of a carbonic anhydrase–Rubisco complex within the alpha-carboxysome. Proc. Natl Acad. Sci. USA 120, e2308600120 (2023).
Yu, W. et al. De novo engineering of programmable and multi-functional biomolecular condensates for controlled biosynthesis. Nat. Commun. 15, 7989 (2024).
Reifenrath, M., Oreb, M., Boles, E. & Tripp, J. Artificial ER-eerived vesicles as synthetic organelles for in vivo compartmentalization of biochemical pathways. ACS Synth. Biol. 9, 2909–2916 (2020).
Einfalt, T. et al. Biomimetic artificial organelles with in vitro and in vivo activity triggered by reduction in microenvironment. Nat. Commun. 9, 1127 (2018).
Mangan, N. M., Flamholz, A., Hood, R. D., Milo, R. & Savage, D. F. pH determines the energetic efficiency of the cyanobacterial CO2 concentrating mechanism. Proc. Natl Acad. Sci. USA 113, E5354–E5362 (2016).
Fei, C., Wilson, A. T., Mangan, N. M., Wingreen, N. S. & Jonikas, M. C. Modelling the pyrenoid-based CO2-concentrating mechanism provides insights into its operating principles and a roadmap for its engineering into crops. Nat. Plants 8, 583–595 (2022).
Rasor, B. J. et al. Toward sustainable, cell-free biomanufacturing. Curr. Opin. Biotechnol. 69, 136–144 (2021).
Opgenorth, P. H., Korman, T. P. & Bowie, J. U. A synthetic biochemistry module for production of bio-based chemicals from glucose. Nat. Chem. Biol. 12, 393–395 (2016).
Trudeau, D. L. et al. Design and in vitro realization of carbon-conserving photorespiration. Proc. Natl Acad. Sci. USA 115, E11455–E11464 (2018).
Opgenorth, P. H., Korman, T. P. & Bowie, J. U. A synthetic biochemistry molecular purge valve module that maintains redox balance. Nat. Commun. 5, 4113 (2014).
Opgenorth, P. H., Korman, T. P., Iancu, L. & Bowie, J. U. A molecular rheostat maintains ATP levels to drive a synthetic biochemistry system. Nat. Chem. Biol. 13, 938–942 (2017).
Yadav, S., Perkins, A. J. P., Liyanagedera, S. B. W., Bougas, A. & Laohakunakorn, N. ATP regeneration from pyruvate in the PURE system. ACS Synth. Biol. 14, 247–256 (2025).
Chowdhury, S. et al. Carbon negative synthesis of amino acids using a cell-free-based biocatalyst. ACS Synth. Biol. 13, 3961–3975 (2024).
Aspacio, D. et al. Shifting redox reaction equilibria on demand using an orthogonal redox cofactor. Nat. Chem. Biol. 20, 1535–1546 (2024).
Berhanu, S., Ueda, T. & Kuruma, Y. Artificial photosynthetic cell producing energy for protein synthesis. Nat. Commun. 10, 1325 (2019).
Lee, K. Y. et al. Photosynthetic artificial organelles sustain and control ATP-dependent reactions in a protocellular system. Nat. Biotechnol. 36, 530–535 (2018).
Miller, T. E. et al. Light-powered CO2 fixation in a chloroplast mimic with natural and synthetic parts. Science 368, 649–654 (2020).
Luo, S. et al. ATP production from electricity with a new-to-nature electrobiological module. Joule 7, 1745–1758 (2023).
Castañeda-Losada, L. et al. Bioelectrocatalytic cofactor regeneration coupled to CO2 fixation in a redox-active hydrogel for stereoselective C–C bond formation. Angew. Chem. Int. Ed. 60, 21056–21061 (2021).
Lavickova, B., Laohakunakorn, N. & Maerkl, S. J. A partially self-regenerating synthetic cell. Nat. Commun. 11, 6340 (2020).
Hagino, K., Masuda, K., Shimizu, Y. & Ichihashi, N. Sustainable regeneration of 20 aminoacyl-tRNA synthetases in a reconstituted system toward self-synthesizing artificial systems. Sci. Adv. 11, eadt6269 (2025).
Schwarz-Schilling, M. et al. Autonomous biogenesis of the entire protein translation machinery excluding ribosomes. Preprint at bioRxiv https://doi.org/10.1101/2024.10.20.619270 (2024).
Ganesh, R. B. & Maerkl, S. J. Towards self-regeneration: exploring the limits of protein synthesis in the protein synthesis using recombinant elements (PURE) cell-free transcription–translation system. ACS Synth. Biol. 13, 2555–2566 (2024).
Giaveri, S. et al. Integrated translation and metabolism in a partially self-synthesizing biochemical network. Science 385, 174–178 (2024).
Nishikawa, S. et al. Amino acid self-regenerating cell-free protein synthesis system that feeds on PLA plastics, CO2, ammonium, and α-ketoglutarate. ACS Catal. 14, 7696–7706 (2024).
Giaveri, S. et al. Nature-inspired circular-economy recycling for proteins: proof of concept. Adv. Mater. 33, 2104581 (2021).
Abil, Z., Giaveri, S., Erb, T. J. & Rothschild, L. J. Integrating metabolism and evolution towards the realization of synthetic life. Nat. Rev. Bioeng. 3, 9–10 (2025).
Tan, X. & Nielsen, J. The integration of bio-catalysis and electrocatalysis to produce fuels and chemicals from carbon dioxide. Chem. Soc. Rev. 51, 4763–4785 (2022).
Beyazay, T. et al. Ambient temperature CO2 fixation to pyruvate and subsequently to citramalate over iron and nickel nanoparticles. Nat. Commun. 14, 570 (2023).
Vornholt, T. et al. Artificial metalloenzymes. Nat. Rev. Methods Primers 4, 78 (2024).
Jack, J., Fu, H., Leininger, A., Hyster, T. K. & Ren, Z. J. Cell-free CO2 valorization to C6 pharmaceutical precursors via a novel electro-enzymatic process. ACS Sustain. Chem. Eng. 10, 4114–4121 (2022).
Landwehr, G. M. et al. A synthetic cell-free pathway for biocatalytic upgrading of one-carbon substrates. Preprint at bioRxiv https://doi.org/10.1101/2024.08.08.607227 (2024).
Dinges, I. et al. Coupling of CO2 electrolysis with parallel and semi‐automated biopolymer synthesis – ex‐cell and without downstream processing. ChemSusChem 17, e202301721 (2024).
Gröger, H., Gallou, F. & Lipshutz, B. H. Where chemocatalysis meets biocatalysis: in water. Chem. Rev. 123, 5262–5296 (2023).
Hu, G. et al. Light-driven CO2 sequestration in Escherichia coli to achieve theoretical yield of chemicals. Nat. Catal. 4, 395–406 (2021).
Guan, X. et al. Maximizing light-driven CO2 and N2 fixation efficiency in quantum dot–bacteria hybrids. Nat. Catal. 5, 1019–1029 (2022).
Liu, J., Zhang, H., Xu, Y., Meng, H. & Zeng, A.-P. Turn air-captured CO2 with methanol into amino acid and pyruvate in an ATP/NAD(P)H-free chemoenzymatic system. Nat. Commun. 14, 2772 (2023).
Waser, V., Mukherjee, M., Tachibana, R., Igareta, N. V. & Ward, T. R. An artificial [Fe4S4]-containing metalloenzyme for the reduction of CO2 to hydrocarbons. J. Am. Chem. Soc. 145, 14823–14830 (2023).
Ding, Y. et al. Nanorg microbial factories: light-driven renewable biochemical synthesis using quantum dot–bacteria nanobiohybrids. J. Am. Chem. Soc. 141, 10272–10282 (2019).
Oehlmann, N. N., Schmidt, F. V., Herzog, M., Goldman, A. L. & Rebelein, J. G. The iron nitrogenase reduces carbon dioxide to formate and methane under physiological conditions: a route to feedstock chemicals. Sci. Adv. 10, eado7729 (2024).
Guo, Y., Hong, X., Chen, Z. & Lv, Y. Electro-enzyme coupling systems for selective reduction of CO2. J. Energy Chem. 80, 140–162 (2023).
Jayathilake, B. S., Bhattacharya, S., Vaidehi, N. & Narayanan, S. R. Efficient and Selective electrochemically driven enzyme-catalyzed reduction of carbon dioxide to formate using formate dehydrogenase and an artificial cofactor. Acc. Chem. Res. 52, 676–685 (2019).
Cheng, C. et al. Electricity-enhanced anaerobic, non-photosynthetic mixotrophy by Clostridium carboxidivorans with increased carbon efficiency and alcohol production. Energy Convers. Manag. 252, 115118 (2022).
Guzman, M. S. et al. Phototrophic extracellular electron uptake is linked to carbon dioxide fixation in the bacterium Rhodopseudomonas palustris. Nat. Commun. 10, 1355 (2019).
Ranaivoarisoa, T. O., Singh, R., Rengasamy, K., Guzman, M. S. & Bose, A. Towards sustainable bioplastic production using the photoautotrophic bacterium Rhodopseudomonas palustris TIE-1. J. Ind. Microbiol. Biotechnol. 46, 1401–1417 (2019).
Bird, L. J. et al. Engineering wired life: synthetic biology for electroactive bacteria. ACS Synth. Biol. 10, 2808–2823 (2021).
Baker, J. J. et al. ML-enhanced peroxisome capacity enables compartmentalization of multienzyme pathway. Nat. Chem. Biol. https://doi.org/10.1038/s41589-024-01759-2 (2024).
Pandi, A. et al. Cell-free biosynthesis combined with deep learning accelerates de novo-development of antimicrobial peptides. Nat. Commun. 14, 7197 (2023).
Landwehr, G. M. et al. Accelerated enzyme engineering by machine-learning guided cell-free expression. Nat. Commun. 16, 865 (2025).
Chainani, Y., Ni, Z., Shebek, K. M., Broadbelt, L. J. & Tyo, K. E. J. DORA-XGB: an improved enzymatic reaction feasibility classifier trained using a novel synthetic data approach. Mol. Syst. Des. Eng. 10, 129–142 (2025).
Acknowledgements
This work was supported by the Max Planck Society, EMBO Postdoctoral Fellowships to B.J.R. (ALTF 337-2023), S.G. (ALTF 162-2022) and A.M.K. (ALTF 684-2022), and a MSCA Fellowship to A.M.K. (Project 101106795 – ECOFix).
Author information
Authors and Affiliations
Contributions
B.J.R., S.G. and A.M.K. wrote the manuscript and designed the figures. T.J.E. guided ideation and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Synthesis thanks Xiulai Chen and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Thomas West, in collaboration with the Nature Synthesis team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rasor, B.J., Giaveri, S., Küffner, A.M. et al. Building complex biochemicals from one-carbon compounds. Nat. Synth 4, 787–798 (2025). https://doi.org/10.1038/s44160-025-00835-2
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s44160-025-00835-2