Abstract
Atherosclerosis underlies most coronary artery disease (CAD). It involves a significant autoimmune component against apolipoprotein B (APOB). In this study, we used short activation-induced marker (AIM) assays to characterize APOB-reactive CD4+ T cells in patients with angiographically verified CAD. APOB-reactive CD4+ T cells expressing CD25 and 4-1BB markers were the most abundant. Their frequency correlated positively with CAD severity. Transcriptomic analysis revealed that these cells were clonally expanded and significantly enriched in genes expressed in tissue-homing effector regulatory T (eTreg) cells. They shared signatures with CD4+ T cells in mouse and human plaques, including expression of the plaque-homing chemokine receptor CXCR6. With increasing disease severity, the Treg signature was progressively and significantly lost. Conversely, APOB-specific Treg cells from patients with severe CAD gained glycolytic and interferon response signatures. We conclude that mild CAD is associated with a regulatory program in APOB-reactive CD4+ T cells, which is replaced by a pro-inflammatory program in patients with severe CAD.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The raw sequencing data are publicly available at the National Center for Biotechnology Informationʼs Gene Expression Omnibus under accession code GSE279783. All other processed data are available in the main text or Supplementary Information. No new material resources were generated in this study. GSEA was done using gene sets in Human ImmuneSigDB (https://www.gsea-msigdb.org/gsea/msigdb/human/genesets.jsp?collection=C7), HALLMARK genes (https://www.gsea-msigdb.org/gsea/msigdb/human/genesets.jsp?collection=H) and from curated T-cell-specific published gene sets (GSE149068, GSE149069, GSE263393 and GSE149090). Human circulating Treg clusters were analyzed using the Broad Institute’s interactive SingleCellPortal (https://singlecell.broadinstitute.org/single_cell/study/SCP1963). Heatmaps were generated based on data in GSE77081 and GSE161426. For analyzing chemokine receptor expression in human coronary plaque T cells, the publicly available dataset (GSE196943) was used.
Code availability
No new algorithms were generated for this study.
References
Libby, P. et al. Atherosclerosis. Nat. Rev. Dis. Primers 5, 56 (2019).
Jonasson, L., Holm, J., Skalli, O., Bondjers, G. & Hansson, G. K. Regional accumulations of T cells, macrophages, and smooth muscle cells in the human atherosclerotic plaque. Arteriosclerosis 6, 131–138 (1986).
Gown, A. M., Tsukada, T. & Ross, R. Human atherosclerosis. II. Immunocytochemical analysis of the cellular composition of human atherosclerotic lesions. Am. J. Pathol. 125, 191–207 (1986).
Roy, P., Orecchioni, M. & Ley, K. How the immune system shapes atherosclerosis: roles of innate and adaptive immunity. Nat. Rev. Immunol. 22, 251–265 (2022).
Fernandez, D. M. et al. Single-cell immune landscape of human atherosclerotic plaques. Nat. Med. 25, 1576–1588 (2019).
Depuydt, M. A. C. et al. Single-cell T cell receptor sequencing of paired human atherosclerotic plaques and blood reveals autoimmune-like features of expanded effector T cells. Nat. Cardiovasc. Res. 2, 112–125 (2023).
Gupta, S. et al. IFN-γ potentiates atherosclerosis in ApoE knock-out mice. J. Clin. Invest. 99, 2752–2761 (1997).
Buono, C. et al. T-bet deficiency reduces atherosclerosis and alters plaque antigen-specific immune responses. Proc. Natl Acad. Sci. USA 102, 1596–1601 (2005).
Buono, C. et al. Influence of interferon-γ on the extent and phenotype of diet-induced atherosclerosis in the LDLR-deficient mouse. Arterioscler. Thromb. Vasc. Biol. 23, 454–460 (2003).
Laurat, E. et al. In vivo downregulation of T helper cell 1 immune responses reduces atherogenesis in apolipoprotein E-knockout mice. Circulation 104, 197–202 (2001).
Ait-Oufella, H. et al. Natural regulatory T cells control the development of atherosclerosis in mice. Nat. Med. 12, 178–180 (2006).
Klingenberg, R. et al. Depletion of FOXP3+ regulatory T cells promotes hypercholesterolemia and atherosclerosis. J. Clin. Invest. 123, 1323–1334 (2013).
Mor, A. et al. Role of naturally occurring CD4+CD25+ regulatory T cells in experimental atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 27, 893–900 (2007).
Mallat, Z. et al. Protective role of interleukin-10 in atherosclerosis. Circ. Res. 85, e17–e24 (1999).
Robertson, A.-K. L. et al. Disruption of TGF-β signaling in T cells accelerates atherosclerosis. J. Clin. Invest. 112, 1342–1350 (2003).
Mor, A., Luboshits, G., Planer, D., Keren, G. & George, J. Altered status of CD4+CD25+ regulatory T cells in patients with acute coronary syndromes. Eur. Heart J. 27, 2530–2537 (2006).
Meng, K. et al. Impairment of circulating CD4+CD25+GARP+ regulatory T cells in patients with acute coronary syndrome. Cell. Physiol. Biochem. 33, 621–632 (2014).
Zhu, Z.-F. et al. Impaired circulating CD4+LAP+ regulatory T cells in patients with acute coronary syndrome and its mechanistic study. PLoS ONE 9, e88775 (2014).
Ma, Y. et al. Imbalanced frequencies of Th17 and Treg cells in acute coronary syndromes are mediated by IL-6-STAT3 signaling. PLoS ONE 8, e72804 (2013).
Han, S. F. et al. The opposite-direction modulation of CD4+CD25+ Tregs and T helper 1 cells in acute coronary syndromes. Clin. Immunol. 124, 90–97 (2007).
Cheng, X. et al. The Th17/Treg imbalance in patients with acute coronary syndrome. Clin. Immunol. 127, 89–97 (2008).
Ammirati, E. et al. Circulating CD4+CD25hiCD127lo regulatory T-cell levels do not reflect the extent or severity of carotid and coronary atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 30, 1832–1841 (2010).
Chowdhury, R. R. et al. Human coronary plaque T cells are clonal and cross-react to virus and self. Circ. Res. 130, 1510–1530 (2022).
Stemme, S. et al. T lymphocytes from human atherosclerotic plaques recognize oxidized low density lipoprotein. Proc. Natl Acad. Sci. USA 92, 3893–3897 (1995).
Caligiuri, G., Paulsson, G., Nicoletti, A., Maseri, A. & Hansson, G. K. Evidence for antigen-driven T-cell response in unstable angina. Circulation 102, 1114–1119 (2000).
Hermansson, A. et al. Inhibition of T cell response to native low-density lipoprotein reduces atherosclerosis. J. Exp. Med. 207, 1081–1093 (2010).
Gistera, A. et al. Low-density lipoprotein-reactive T cells regulate plasma cholesterol levels and development of atherosclerosis in humanized hypercholesterolemic mice. Circulation 138, 2513–2526 (2018).
Nilsson, J., Bjorkbacka, H. & Fredrikson, G. N. Apolipoprotein B100 autoimmunity and atherosclerosis—disease mechanisms and therapeutic potential. Curr. Opin. Lipidol. 23, 422–428 (2012).
Khan, A., Roy, P. & Ley, K. Breaking tolerance: the autoimmune aspect of atherosclerosis. Nat. Rev. Immunol. 24, 670–679 (2024).
Roy, P. et al. Immunodominant MHC-II (major histocompatibility complex II) restricted epitopes in human apolipoprotein B. Circ. Res. 131, 258–276 (2022).
Wolf, D. et al. Pathogenic autoimmunity in atherosclerosis evolves from initially protective apolipoprotein B100-reactive CD4+ T-regulatory cells. Circulation 142, 1279–1293 (2020).
Kimura, T. et al. Regulatory CD4+ T cells recognize MHC-II-restricted peptide epitopes of apolipoprotein B. Circulation 138, 1130–1143 (2018).
Saigusa, R. et al. Single cell transcriptomics and TCR reconstruction reveal CD4 T cell response to MHC-II-restricted APOB epitope in human cardiovascular disease. Nat. Cardiovasc. Res. 1, 462–475 (2022).
Bacher, P. et al. Regulatory T cell specificity directs tolerance versus allergy against aeroantigens in humans. Cell 167, 1067–1078 (2016).
Meckiff, B. J. et al. Imbalance of regulatory and cytotoxic SARS-CoV-2-reactive CD4+ T cells in COVID-19. Cell 183, 1340–1353 (2020).
Renand, A. et al. Integrative molecular profiling of autoreactive CD4 T cells in autoimmune hepatitis. J. Hepatol. 73, 1379–1390 (2020).
Reiss, S. et al. Comparative analysis of activation induced marker (AIM) assays for sensitive identification of antigen-specific CD4 T cells. PLoS ONE 12, e0186998 (2017).
Lemieux, A. et al. Enhanced detection of antigen-specific T cells by a multiplexed AIM assay. Cell Rep. Methods 4, 100690 (2024).
Ammirati, E. et al. Effector memory T cells are associated with atherosclerosis in humans and animal models. J. Am. Heart Assoc. 1, 27–41 (2012).
Rattik, S. et al. Elevated circulating effector memory T cells but similar levels of regulatory T cells in patients with type 2 diabetes mellitus and cardiovascular disease. Diab. Vasc. Dis. Res. 16, 270–280 (2019).
Nettersheim, F. S. et al. PD-1 and CD73 on naive CD4+ T cells synergistically limit responses to self. Nat. Immunol. 26, 105–115 (2025).
Schmiedel, B. J. et al. Impact of genetic polymorphisms on human immune cell gene expression. Cell 175, 1701–1715 (2018).
Höllbacher, B. et al. Transcriptomic profiling of human effector and regulatory T cell subsets identifies predictive population signatures. Immunohorizons 4, 585–596 (2020).
Attaf, M., Huseby, E. & Sewell, A. K. αβ T cell receptors as predictors of health and disease. Cell Mol. Immunol. 12, 391–399 (2015).
Roy, P., Suthahar, S. S. A., Makings, J. & Ley, K. Identification of apolipoprotein B–reactive CDR3 motifs allows tracking of atherosclerosis-related memory CD4+T cells in multiple donors. Front. Immunol. 15, 1302031 (2024).
Ding, Y., Xu, J. & Bromberg, J. S. Regulatory T cell migration during an immune response. Trends Immunol. 33, 174–180 (2012).
Lee, J. H., Kang, S. G. & Kim, C. H. FoxP3+ T cells undergo conventional first switch to lymphoid tissue homing receptors in thymus but accelerated second switch to nonlymphoid tissue homing receptors in secondary lymphoid tissues. J. Immunol. 178, 301–311 (2007).
Iellem, A. et al. Unique chemotactic response profile and specific expression of chemokine receptors CCR4 and CCR8 by CD4+CD25+ regulatory T cells. J. Exp. Med. 194, 847–853 (2001).
Delacher, M. et al. Single-cell chromatin accessibility landscape identifies tissue repair program in human regulatory T cells. Immunity 54, 702–720 (2021).
Yasumizu, Y. et al. Single-cell transcriptome landscape of circulating CD4+ T cell populations in autoimmune diseases. Cell Genom. 4, 100473 (2024).
Huehn, J. & Hamann, A. Homing to suppress: address codes for Treg migration. Trends Immunol. 26, 632–636 (2005).
Li, J. et al. CCR5+T-bet+FoxP3+ effector CD4 T cells drive atherosclerosis. Circ. Res. 118, 1540–1552 (2016).
Galkina, E. et al. CXCR6 promotes atherosclerosis by supporting T-cell homing, interferon-γ production, and macrophage accumulation in the aortic wall. Circulation 116, 1801–1811 (2007).
Proudfoot, A. E. I. Chemokine receptors: multifaceted therapeutic targets. Nat. Rev. Immunol. 2, 106–115 (2002).
Freuchet, A. et al. Identification of human exTreg cells as CD16+CD56+ cytotoxic CD4+ T cells. Nat. Immunol. 24, 1748–1761 (2023).
Wang, Z. et al. Pairing of single-cell RNA analysis and T cell antigen receptor profiling indicates breakdown of T cell tolerance checkpoints in atherosclerosis. Nat. Cardiovasc. Res. 2, 290–306 (2023).
Cretney, E., Kallies, A. & Nutt, S. L. Differentiation and function of Foxp3+ effector regulatory T cells. Trends Immunol. 34, 74–80 (2013).
Huehn, J. et al. Developmental stage, phenotype, and migration distinguish naive- and effector/memory-like CD4+ regulatory T cells. J. Exp. Med. 199, 303–313 (2004).
Luo, Y. et al. Single-cell transcriptomic analysis reveals disparate effector differentiation pathways in human Treg compartment. Nat. Commun. 12, 3913 (2021).
Mijnheer, G. et al. Conserved human effector Treg cell transcriptomic and epigenetic signature in arthritic joint inflammation. Nat. Commun. 12, 2710 (2021).
Kumagai, S. et al. The PD-1 expression balance between effector and regulatory T cells predicts the clinical efficacy of PD-1 blockade therapies. Nat. Immunol. 21, 1346–1358 (2020).
Kamada, T. et al. PD-1+ regulatory T cells amplified by PD-1 blockade promote hyperprogression of cancer. Proc. Natl Acad. Sci. USA 116, 9999–10008 (2019).
Tan, C. L. et al. PD-1 restraint of regulatory T cell suppressive activity is critical for immune tolerance. J. Exp. Med. 218, e20182232 (2021).
Campbell, D. J. & Koch, M. A. Phenotypical and functional specialization of FOXP3+ regulatory T cells. Nat. Rev. Immunol. 11, 119–130 (2011).
Zhang, Z. et al. Activation and functional specialization of regulatory T cells lead to the generation of Foxp3 instability. J. Immunol. 198, 2612–2625 (2017).
Schmiedel, B. J. et al. Single-cell eQTL analysis of activated T cell subsets reveals activation and cell type–dependent effects of disease-risk variants. Sci. Immunol. 7, eabm2508 (2022).
Zeng, H. & Chi, H. Metabolic control of regulatory T cell development and function. Trends Immunol. 36, 3–12 (2015).
Zhao, T. X., Newland, S. A. & Mallat, Z. 2019 ATVB Plenary Lecture: Interleukin-2 therapy in cardiovascular disease: the potential to regulate innate and adaptive immunity. Arterioscler. Thromb. Vasc. Biol. 40, 853–864 (2020).
Vigario, F. L., Kuiper, J. & Slutter, B. Tolerogenic vaccines for the treatment of cardiovascular diseases. eBioMedicine 57, 102827 (2020).
Zhao, T. X. et al. Regulatory T-cell response to low-dose interleukin-2 in ischemic heart disease. NEJM Evid. 1, EVIDoa2100009 (2022).
Dikiy, S. & Rudensky, A. Y. Principles of regulatory T cell function. Immunity 56, 240–255 (2023).
Loffredo, L. F., Savage, T. M., Ringham, O. R. & Arpaia, N. Treg–tissue cell interactions in repair and regeneration. J. Exp. Med. 221, e20231244 (2024).
Raposo-Gutiérrez, I., Rodríguez-Ronchel, A. & Ramiro, A. R. Atherosclerosis antigens as targets for immunotherapy. Nat. Cardiovasc. Res. 2, 1129–1147 (2023).
Acknowledgements
We thank members of the Clinical Core, Flow Cytometry Core and Sequencing Core at the La Jolla Institute for Immunology. We thank members of the cardiac catheterization laboratories at the University of Virginia. We thank the Augusta University Georgia Cancer Center Flow and Mass Cytometry Core Facility (RRID: SCR_025747). We acknowledge support from the National Institutes of Health (awards P01 HL136275 and R35 HL145241) to K.L.
Author information
Authors and Affiliations
Contributions
K.L. and P.R. designed the study. P.R. conducted most experiments and analyzed and assembled data. A.B. and S.S.A.S. conducted most of the computational data processing. A.B., S.S.A.S., M.O. and M.M. analyzed data. Q.L., S.P. and S.K. conducted flow cytometry experiments. J.M. performed TCR extraction and analysis. R.W. prepared Smart-seq2 libraries. A.W.T.C. provided collaborative input for gene expression analyses. A.S. provided critical expertise and supportive data for stimulation assays. C.A.M. provided clinical expertise, patient samples and medical data. P.R., A.B. and S.S.A.S. prepared the figures. K.L. acquired funds. K.L. and P.R. supervised the study and wrote the paper. All authors discussed the data and provided critical input.
Corresponding authors
Ethics declarations
Competing interests
K.L. is the founder and co-owner of Atherovax, Inc. He receives no compensation from Atherovax. No Atherovax funds were used in this study. K.L. and P.R. are named as co-inventors on a patent application (provisional application no. 63/789,764, filed by the La Jolla Institute for Immunology, approval status pending) that is related to the use of human APOB epitopes and related methods in modulating inflammatory responses and treating adverse cardiovascular events, disease and atherosclerosis. The other authors declare no competing interests.
Peer review
Peer review information
Nature Cardiovascular Research thanks Claudia Monaco and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
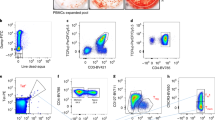
Extended Data Fig. 1 Flow cytometry-based evaluation of activation marker expression in APOB-reactive human CD4+T cells.
a-b) Human PBMCs (n = 18 independent donors) were stimulated with APOB6 peptide pool and expression of different activation-induced marker (AIM) combinations in stimulated vs unstimulated PBMCs were compared. a) Mean ± SEM yield per million CD4+T cells (numbers in stimulated minus those in unstimulated sets) for nine AIM combinations are shown. b) Average (mean ± SEM) fold change (frequency in stimulated divided by that in unstimulated) for CD40L + CD69 + , CD25 + 4-1BB + , CD25 + OX-40+ and CD69 + OX-40 + AIM combinations are shown. c-e) Analysis of APOB-reactive CD4+T cells in CAD patients (n = 18) related to Fig. 1. c) Gating strategy in flow cytometry to identify AIM1,2,3 and AIM- subsets shown in Fig. 1a. d) Pie-chart showing relative abundance of each APOB-reactive CD4+T subset (AIM1, 2 and 3) within total AIM+ cells. Average frequencies were plotted to calculate percent abundance. e) Frequencies (mean ± SEM) of AIM1+ (left) and AIM3+ (right) cells in CADlo (Gensini<20, n = 9, mean Gensini score 10.6; SD ± 8.3) vs CADhi groups (Gensini>20, n = 9, mean Gensini 70.9; SD ± 28.7). Log10 transformed Y-axes; data points with 0 or negative values were collapsed onto the minimum value on the scale. Statistical tests done with two-tailed Mann-Whitney U test (e).
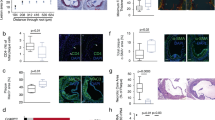
Extended Data Fig. 2 Gene expression in APOB-reactive and control transcriptomes from CAD patients.
a) Clinical table summarizing details related to demographics, medications, lab values and disease severity of patients in the CAD cohort used for transcriptomic analyses. Categorical variables are plotted as counts and percentages, while continuous variables are shown as mean ± SEM within the cohort (sample size = 40 patients). b) Table showing donor IDs (as used in the CAVA cohort), CAD severity scores (Gensini), and input cell numbers for cDNA preparation for each APOB-reactive (AIM1,2,3) and control (AIM-) libraries that were sequenced. Samples labelled as “Dropout” were excluded due to poor quality and yield of cDNA or library. All samples from donor #472 were excluded due to low ( < 50%) viability of PBMCs from this donor. Median viability of PBMCs from other donors was 93.1% (Min-max 80.34 – 98.1, interquartile range 4.7). c) Venn diagram depicting overlap across genes downregulated in AIM1, 2 and 3 subsets as compared to the AIM- group.
Extended Data Fig. 3 Analysis of gene expression across three APOB-reactive subsets.
a) Elbow plot analysis to identify the number of PCs that contribute to >90% of the variance in the transcriptomes. b) Volcano plots showing number of DE genes between AIM2 and AIM1 (left), AIM3 and AIM1 (middle), AIM3 and AIM2 (right). Y axis capped at p = 10-14. Horizontal line at −log10 (p-value) = 1.3 (representing adjusted p-value 0.05). Vertical lines at log2fold ± 1. Statistical analyses were performed using a two-tailed Wald test with Benjamini–Hochberg p-value adjustment. Criteria for significant DE: log2fold ± 1, adjusted p-value < 0.05. Red: upregulated; Blue: downregulated; Grey dots: not significantly different. c) Heatmap of APOB-enriched genes ranked based on their enrichment in AIM1 (top 50) or AIM2&3 (bottom 50) transcriptomes.
Extended Data Fig. 4 Flow cytometry analysis of Treg-related markers in three APOB-reactive subsets.
a) Gating strategy in flow cytometry to identify AIM1,2,3 and AIM- subsets. b-d) Representative FACS plots showing FOXP3 (b), HELIOS (c) and CTLA4 (d) protein expression in AIM1, AIM2, AIM3 and AIM- subsets.
Extended Data Fig. 5 Clonality analysis of APOB-reactive TCRβ CDR3 repertoire.
Table showing the number of expanded CDR3 clones from APOB-reactive subsets (AIM1-3 and total AIM + ) within each donor. The final column shows the number of AIM+ clones that were also detected in the control AIM- CDR3 repertoire from the same donor.
Extended Data Fig. 6 Chemokine receptor expression in APOB-reactive Tregs and all Tregs.
a) Gating strategy in flow cytometry to identify AIM2+ Tregs, total Tregs and Non-APOB (AIM-) subsets. b) Representative FACS plots (left) and quantification (mean ± SEM, right) showing expression of CCR5 protein marker on APOB-reactive Treg subset (green) and on all CD25+CD127lo Treg cells (black) in human PBMCs (n = 6). Non-APOB reactive (AIM-) cells and Fluorescence minus one (FMO) were used as negative controls. Statistical comparisons (b) were done using two-tailed Mann–Whitney U test.
Extended Data Fig. 7 APOB-reactive Tregs are enriched in effector Treg markers.
a) Median expression levels (TPMs) of general and effector Treg-related genes in APOB-reactive Tconv (AIM1, n = 37), Treg (AIM2, n = 35), and non-APOB reactive control (AIM-, n = 38) samples. b-d) Gene expression analysis in circulating human Treg subsets accessed at Broad Institute’s SingleCellPortal (SCP1963). UMAP visualization showing CCR4, CCR8, CXCR6 (b) and TIGIT (c) in Treg subsets. Color scale: yellow (lowest) to blue (highest) expression. Red outline marks FOXP3 expressing Treg cells. d) Violin plots of HLA-DRA, HLA-DRB1, and HLA-DRB5 genes (n = 13 subjects). e) FACS plots showing surface expression of TIGIT (left) and HLA-DR (right) proteins on APOB-reactive Treg subset and on all CD25+CD127lo Treg cells in human PBMCs. f) Significantly enriched pathways related to effector Treg genes described in Fig. 6g. Dotted line at -log10 adjusted p = 1.3. g) FACS plots showing FMO control for PD-1 expression on human AIM2 (left) and total Treg (right) cells. Gating strategy to identify AIM2+ Tregs and total Tregs (e,g) shown in ED Fig. 6a. Statistical comparisons were done using the Kruskal-Wallis test with Dunn’s adjustment (a) and two-tailed Fisher’s exact test and Benjamini-Hochberg adjustment (f).
Extended Data Fig. 8 Dynamics of Treg gene expression in APOB-reactive Treg transcriptomes from patients with varying CAD severity.
a) GSEA plot showing negative enrichment of activated Treg signature in APOB-reactive Treg transcriptomes from patients with highest CAD severity (Gensini >30) compared to those with less severe CAD (Gensini <30). Enrichment score was calculated using a weighted Kolmogorov–Smirnov-like statistic and phenotype-based permutation test in-built in GSEA. b) Heatmaps showing expression (median TPMs) of Treg genes from DICE in AIM2 transcriptomes from donors grouped by CAD severity. c) Table showing NES values for Th1/Th17 signatures in APOB-reactive Treg transcriptomes from CAD patients with Gensini 10-20, 20.5-30 and >30.
Supplementary information
Supplementary Tables 1–7
Supplementary Table 1: Class II HLA alleles of CAVA donors used for transcriptomic analyses. Supplementary Table 2: Gene sets used for signature analysis. Supplementary Table 3: Distinct and overlapping sets of genes differentially expressed between AIM− and each AIM+ subset. Supplementary Table 4: Genes differentially expressed across the three APOB-reactive AIM+ groups. Supplementary Table 5: Productive (in-frame for protein translation and without stop codons) TCR β chain CDR3 clones. Supplementary Table 6: Normalized expression values of antigen-specific and eTreg-related genes. Supplementary Table 7: Normalized gene expression values of memory Treg genes.
Source data
Source Data Fig. 1
Numerical source data.
Source Data Fig. 2
Numerical source data.
Source Data Fig. 3
Numerical source data.
Source Data Fig. 4
Numerical source data.
Source Data Fig. 5
Numerical source data.
Source Data Fig. 6
Numerical source data.
Source Data Fig. 7
Numerical source data.
Source Data Extended Data Fig./Table 1
Numerical source data.
Source Data Extended Data Fig./Table 6
Numerical source data.
Source Data Extended Data Fig./Table 7
Numerical source data.
Source Data Extended Data Fig./Table 8
Numerical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Roy, P., Bellapu, A., Suthahar, S.S.A. et al. Loss of effector Treg signature in APOB-reactive CD4+ T cells in patients with coronary artery disease. Nat Cardiovasc Res 4, 841–856 (2025). https://doi.org/10.1038/s44161-025-00671-9
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s44161-025-00671-9