Abstract
Alzheimer’s disease (AD) is a chronic, incurable brain disorder, and early detection is essential for effective management. Traditional detection methods often rely on large, pre-labeled image datasets, which are costly and difficult to compile. To address this, we propose an innovative active learning framework that improves model performance using fewer labeled samples. Conventional active learning techniques often use static selection strategies that lack adaptability. To address this, the method combines deep reinforcement learning (DRL) with a scope loss function (SLF) to improve flexibility. This allows a dynamic balance between exploiting known data and exploring new data opportunities. To reduce hyperparameter sensitivity in DRL, we apply an advanced differential evolution (DE) algorithm. The model was evaluated on the OASIS and ADNI datasets, achieving F-measures of 92.044% and 93.685%, showing its superiority in early AD detection.
Similar content being viewed by others
Introduction
Globally, around 55 million persons suffered from dementia in 2021, a number that is expected to rise to 78 million and 139 million by 2030 and 2050, respectively1,2. A primary cause of dementia, Alzheimer’s disease (AD) is a neurodegenerative disease characterized by neuron loss and the buildup of amyloid plaques and neurofibrillary tangles in the brain. It causes memory and cognitive decline, with a devastating impact on mental and social functioning. A definitive diagnosis of AD is confirmed via invasive brain biopsy (usually post-mortem). Still, the diagnosis can be presumed based on clinical assessment and advanced neuroimaging such as computed tomography (CT), positron emission tomography (PET), and magnetic resonance imaging (MRI), including functional MRI3,4. MRI is the diagnostic modality of choice. It utilizes non-ionizing static and dynamic magnetic fields to produce high-resolution images depicting detailed brain structure, including tissue atrophy and ventricular enlargement, which are common in AD5, with high tissue contrast. MRI can detect early-stage abnormalities in AD and is useful for monitoring disease progression and therapeutic efficacy6.
Artificial intelligence has advanced many computer-aided fields7,8,9, including automated AD detection10,11. Several traditional machine learning models have been published12,13,14,15,16,17,18, but they are often inadequate for handling high-dimensional and inherently heterogeneous MRI data. Typically requiring handcrafted pre-processing and feature engineering, these models may still fail to capture nonlinear interactions and patterns within the data. Deep learning, especially convolutional neural networks (CNNs), has improved automatic feature extraction in AD diagnosis19,20,21,22,23. However, CNNs demand large annotated datasets, which are costly and impractical to obtain in medical settings24. This motivates the need for strategies that reduce labeling effort while preserving model accuracy25.
Active learning offers a promising approach by selecting the most informative unlabeled samples for manual annotation and model training26. However, most traditional active learning methods rely on handcrafted heuristics27, such as uncertainty or representativeness scores28, which are static and do not adapt to the learning progression of the model. These methods treat all training stages equally, regardless of how the internal knowledge changes of the model over time. For example, a sample considered “uncertain” early in training may become confidently predictable later, but static strategies cannot recognize such shifts. This lack of adaptability leads to inefficient sample selection, redundant labeling, and slower convergence, particularly in deep CNN-based medical imaging tasks where model behavior evolves significantly during training.
Deep reinforcement learning (DRL) offers a flexible approach to active learning, where the method for selecting samples evolves as the model learns. In contrast to traditional methods that employ fixed rules, DRL utilizes feedback from training to evaluate the value of labeling each sample and adjusts its selection process over time. For instance, it learns to skip samples that were initially uncertain but later become predictable, thereby avoiding redundant labeling. This dynamic behavior enables the model to focus on the most informative data at each stage, making the labeling process more efficient and accelerating convergence. Although DRL has been applied for detecting AD using MRI29,30,31, to the best of our knowledge, no studies have yet explored DRL within an active learning framework for AD. However, DRL approaches are limited by the exploitation-exploration dilemma and the high sensitivity to hyperparameter settings32.
An optimal balance between exploitation and exploration is important to avoid settling on a suboptimal strategy that outweighs immediate benefits. To this end, considerable research has focused on incorporating a scope loss function (SLF) to optimize the fine-tuning of the balance between exploiting known data to gain more confidence in predictions and exploring new data to uncover novel insights33. SLF indirectly promotes exploration by limiting how quickly an agent becomes overly exploitative. It also ensures that gradients do not excessively favor high-certainty actions. By dynamically adjusting gradient magnitudes relative to action certainty, SLF allows agents to maintain a broader search space and better avoid convergence to suboptimal policies.
Several methods have been proposed to reduce hyperparameter sensitivity in DRL, including grid search and genetic algorithms34. Grid search systematically assesses all hyperparameter combinations within a structured framework to pinpoint the optimal settings, which is a resource-intensive process. In contrast, the genetic algorithm emulates evolutionary processes to refine hyperparameters incrementally toward a more effective configuration, which is more efficient but risks convergence issues and settling for suboptimal solutions. Differential evolution (DE) algorithms35 overcome these limitations by employing differential vectors to refresh the current solution pool and achieve global optima without hyperparameter discretization. DE algorithms possess three phases: mutation, crossover, and selection. During mutation, new solutions are generated by proportionally adjusting differences among existing solutions. During crossover, mutated vectors are merged with existing solutions. During the selection process, created solutions are evaluated against existing ones, and the most effective are retained. Genetic diversity is enhanced, preventing stagnation of the optimization process and rendering the algorithm adaptable and efficient in navigating the complex solution space36.
Previous studies have either used static active learning heuristics37,38 or applied DRL separately39,40 for AD detection. To the best of our knowledge, this is the first study to combine DRL-based active learning with scope loss and DE within a single unified framework. This approach addresses the dual challenge of adaptive sample selection and robust model optimization, enhancing training efficiency while reducing annotation costs. This highlights the novelty of our work in integrating adaptive sample selection with efficient hyperparameter tuning. Contributions of the proposed model are as follows:
-
DRL facilitates active learning by dynamically selecting the most informative unlabeled images for labeling. It increases learning efficiency (and lowers training costs) through continuous adaptation of the training set, ensuring the model remains relevant and updated with the most valuable data throughout the training.
-
SLF balances exploiting known data to gain more predictive confidence versus exploring new data to uncover novel insights, enabling the model to make more accurate decisions in complex scenarios.
-
Enhanced DE systematically fine-tunes model hyperparameters to specific datasets and task characteristics, ensuring optimal and efficient model operations.
-
Training and testing our model on public AD MRI datasets, we achieved an average F1-measure of over 90% for differentiating between normal and AD images.
The rest of the paper is organized as follows. Section “Methods” details our model and methods; Section “Results” presents the experimental results; Section “Discussion” discusses and proposes future research directions.
Methods
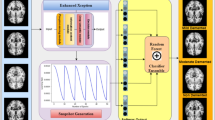
Figure 1 illustrates the overall architecture of the proposed model for AD detection using an active learning framework based on DRL. The model consists of three main components: the classifier, the Q-network, and the hyperparameter optimization module.
-
Input data block: The model starts with two sets of data: a small batch of labeled MRI scans (classified as Alzheimer’s or Non-Alzheimer’s) and a large pool of unlabeled samples. To improve the detection of AD, the model utilizes active learning to efficiently identify useful training data. The selected samples are added to the training set, enabling the classifier to retrain on more informative data and improve its diagnostic accuracy.
-
Classifier module: This component includes multiple CNNs that extract features from labeled MRI scans. The extracted features (\(F\)) are input into a fully connected (FC) layer to perform the final classification. The classifier is trained and updated throughout the process using a focal loss (FL) to handle class imbalance.
-
Q-Network module: The Q-network evaluates unlabeled samples and decides whether each one should be annotated. It receives feature information from the CNNs in the classifier module and updates its policy based on classification outcomes. Scope loss regulates the trade-off between exploration (labeling new, diverse samples) and exploitation (focusing on familiar informative patterns). This prevents the model from converging prematurely to suboptimal policies.
-
Active learning loop: Once the Q-network selects a new batch of samples, they are labeled and added to the annotated dataset. The classifier is retrained using this updated data, and the Q-network adjusts its strategy accordingly. This iterative loop continues until the labeling budget is used up or the performance of the model stabilizes.
-
Hyperparameter optimization by DE: The classifier and Q-network are fine-tuned using DE algorithms. A random key encoding combined with k-means clustering selects mutation candidates from the population group with the lowest average objective value. This strategy accelerates convergence and improves global optimization.
Framework
We analyze the image dataset \(X={\{{x}_{i}\}}_{i=1}^{N}\)N, where \({x}_{i}\) represents the \(i\)-th image in the dataset, and \(N\) is the total number of training images. These images are categorized into classes \(c=\{{c}_{i}\}\), where \({c}_{i}\) represents one of the \(C\) classes. Labeled images in the dataset are \({X}_{L}={\{({x}_{{L}_{i}},{y}_{{L}_{i}})\}}_{i=1}^{{N}_{L}}\), where \({x}_{{L}_{i}}\) represents the i-th labeled image; \({y}_{{L}_{i}}\), its class label; and \({N}_{L}\), number of labeled images. Unlabeled images in the dataset are \({X}_{U}={\{{x}_{{U}_{i}}\}}_{i=1}^{{N}_{U}}\), where \({x}_{{U}_{i}}\) represents the i-th unlabeled image; and \({N}_{U}\), number of unlabeled images. The DRL model is structured as follows:
-
State. At each decision point, the agent evaluates the current context, which must contain critical and pertinent information. The state is characterized by combining the feature vectors from the CNNs, \({f}_{i}\), with the predicted scores \(f({x}_{i})\). These states are represented as \(S=\{{s}_{i}^{t}\}\), where \({s}_{i}^{t}=({f}_{i}^{t},f({x}_{i}^{t}))\); and t, the specific time point.
-
Action. Actions decide if a human should label a sample. The range of options is represented as \(A=\{\mathrm{0,1}\}\). When \({a}_{i}^{t}=1\), it assigns \({x}_{i}^{t}\) for labeling by a human, who then annotates the sample, which is then incorporated into the labeled dataset. Active learning ends when either all samples have been used up, or the labeling budget is depleted.
-
Reward. While many methods delay rewards until the end of a sequence, our approach provides interim rewards to link each decision directly with its long-term effects. The reward structure is in two parts:
-
1.
For actions that trigger annotation (\({a}_{i}^{t}=1\)), the reward function applies a minor penalty of −0.2, \(R\left({s}_{i}^{t-1},{a}_{i}^{t}\right)=-0.2\), to discourage excessive annotation and conserve resources.
-
2.
For actions that avoid annotation (\({a}_{i}^{t}=0\)), the entropy loss function of unlabeled samples is calculated using centroids of feature vectors. \({m}_{c}\), the centroid of the feature vector for class \(c\), is given by the following formula:
$${m}_{c}=\frac{1}{{n}_{c}}\mathop{\sum}\limits _{i\in{\pi }_{c}}{f}_{i}$$(1)
where \({n}_{c}={|\pi }_{c}|\) which sums up sample indices belonging to class \(c\). The entropy loss function for the unlabeled samples is calculated as:
$$L\left({x}_{{U}_{i}},c\right)=-\mathop{\sum }\nolimits_{j=1}^{C}\frac{{e}^{-{\left({f}_{{U}_{i}}-{m}_{{c}_{j}}\right)}^{2}}}{\mathop{\sum }\nolimits_{j=1}^{C}{e}^{-{\left({f}_{{U}_{i}}-{m}_{{c}_{j}}\right)}^{2}}}\log \frac{{e}^{-{\left({f}_{{U}_{i}}-{m}_{{c}_{j}}\right)}^{2}}}{\mathop{\sum }\nolimits_{j=1}^{C}{e}^{-{\left({f}_{{U}_{i}}-{m}_{{c}_{j}}\right)}^{2}}}$$(2)where \({f}_{{U}_{i}}\) represents the feature vector for the i-th unlabeled image extracted by the CNNs. The reward function for when \({a}_{i}^{t}=0\) is defined as:
$$R\left({s}_{i}^{t-1},{a}_{i}^{t}\right)=\left\{\begin{array}{c}1,L({x}_{{U}_{i}},c) < \partial \\ -1,L({x}_{{U}_{i}},c) > \partial \end{array}\right.$$(3)where \(L({x}_{{U}_{i}},\,c)\) measures the entropy loss for an unlabeled image \({x}_{{U}_{i}}\), with \(\partial\) an adjustable parameter that shifts dynamically based on the model’s evolving accuracy. This reward structure encourages the model to label samples that it classifies with low confidence, moving beyond conventional uncertainty-based methods. Initially, \(\partial\) is set lower to reflect the preliminary unreliability of the extracted features. As training advances and the dataset of annotated samples expands, \(\partial\) gradually increases, signaling enhanced reliability in the feature extraction process. This progressive method ensures that as the model’s accuracy improves, it increasingly relies on its refined judgment to request annotations for only the most essential samples, thereby maximizing the efficiency of labeling resources.
-
1.
Our model utilizes a series of CNNs to perform feature extraction. Each CNN comprises five two-dimensional (2D) convolutional layers with filters progressively reducing in size from 128 to 8, tailored to enhance the granularity of feature analysis throughout the image processing stages. Each convolutional layer is equipped with uniform 2\(\times\)2 kernels, a stride of 3, and padding of 4, ensuring an efficient feature extraction without losing important image details. After the convolutional stages, a 2\(\times\)2 max-pooling layer reduces the spatial dimensions of the feature maps, effectively retaining crucial features while boosting the processing speed. The outputs of all CNNs are amalgamated into a single vector, designated as vector \(F\) (Fig. 1). This vector diverges into two distinct pathways: the Q-network and the classification channels. The Q-network, integral to the model’s decision-making process, includes two FC layers. The classification pathway outputs a normalized FC layer value, \(f({x}_{i})\), while the Q-network produces a 2D vector representing potential actions (\({a}_{i}=0\) or \({a}_{i}=1\)). The reinforcement learning agent then chooses the action corresponding to the higher value indicated by the Q-network’s output.
For the classifier training, we utilize FL41 to address the issue of imbalanced classification. FL modifies the standard cross-entropy loss to focus more on hard, misclassified examples while reducing the relative loss for well-classified instances. This is particularly effective in scenarios with a significant class imbalance, as it prevents the overwhelming number of easy negatives from dominating the gradient. FL achieves this by introducing a modulating factor to the cross-entropy loss, which adds a weighting factor to the standard loss, dynamically adjusting the rate at which easy examples are learned. This allows the model to focus more on challenging examples with a higher likelihood of being from the minority class, thereby promoting more balanced learning and preventing the common issue of the majority class overshadowing the minority class’s influence. Using FL, our model can enhance feature learning from underrepresented classes, leading to more equitable and accurate classification performance across diverse classes.
The training of the Q-network initiates with the specification of a transition tuple \(({s}_{i}^{t},{a}_{i}^{t},{r}_{i}^{t},{s}_{i}^{t+1})\), encapsulating an event in the DRL agent’s journey. This tuple details the process where the agent, upon executing action \({a}_{i}^{t}\) in state \({s}_{i}^{t}\), garners a reward \({r}_{i}^{t}\) and progresses to the subsequent state \({s}_{i}^{t+1}\). The Q-network plays a critical role in selecting high-quality samples for annotation. This selection process has a direct impact on the accuracy of the classifier. When the Q-network selects samples that add little new information or resemble existing labeled data, the classifier gains minimal benefit. This may slow down progress or reduce performance. To mitigate such risks, we utilize a scope loss function (SLF) that discourages overly confident actions and promotes the exploration of diverse data regions. Furthermore, the Q-network is periodically updated based on feedback from the classifier. This helps correct early-stage biases and better align sample selection with the classifier’s evolving learning needs.
Scope loss
In DRL, striking the optimal balance between exploration and exploitation is essential. DRL agents continuously adapt their strategies by interacting with and learning from their environment, significantly influencing their decision-making process. These agents are tasked with exploring and discovering actions that might yield high rewards, which is crucial for thorough learning. Concurrently, they must exploit their existing knowledge to optimize their gains. Initially, agents might prioritize exploration, but as they grow more acquainted with the environment, the emphasis gradually shifts toward exploitation. Overemphasis on exploration can hinder the development of effective strategies, while excessive exploitation might prevent the discovery of potentially superior options42.
During the training of our model, we introduce a sophisticated loss function called SLF. This loss function, which incorporates elements from focal loss, policy loss, and entropy loss, plays a pivotal role. Like FL, SLF addresses class imbalance by adjusting the gradients for each class based on their prediction certainty. It also encourages exploration, similar to policy and entropy losses, by penalizing certain policies. As shown in Eq. 4, in the actor-critic architecture, the SLF loss uses \(A-{{\rm{\alpha }}p}_{i}\) as a scaling factor, where \(\alpha\) is a hyperparameter that adjusts \({p}_{i}\) relative to the magnitude of \(A\). This formula fosters a balanced learning dynamic, enhancing both the accuracy and exploratory actions within the model’s training33.
where policy and entropy losses broadly incentivize exploration by indiscriminately introducing explorative elements into the agent’s behavior, SLF uniquely promotes exploration by moderating how quickly an agent can adopt exploitative tactics. With SLF, a positive value of \(A\) means that higher confidence (\({p}_{i}\)) results in smaller gradient adjustments, thus delaying premature exploitation. It is crucial to mention that SLF can be simplified by removing the bias term from its formula, potentially allowing the agent to reach an optimal policy more effectively. Moreover, SLF can be redefined by setting \(\gamma =1\) in its core equation, thereby eliminating the dependence on additional hyperparameters. In scenarios of supervised classification, setting both \(A\) and \(\alpha\) to 1 streamlines the implementation, as illustrated in Eq. 533.
Setting the ideal \(\alpha\) value in DRL presents a challenge because the distribution of the advantage term, \(A\), changes across different settings and as training progresses. The chosen \(\alpha\) must be small enough to let the advantage term predominantly dictate the scope adjustment. Instead of manually tuning \(\alpha\) for each specific environment, employing a consistent \(\alpha\) alongside z-score batch normalization of the advantages, as depicted in Eq. 6, establishes a universally optimal \(\alpha\) that is effective across various settings and throughout training33.
Scope loss for proximal policy optimization
We have integrated SLF into actor-critic DRL algorithms and supervised classification frameworks. Considering the effectiveness of proximal policy optimization (PPO) in distributed systems, we are further adapting SLF loss for use in PPO architectures. This adaptation begins with the conventional policy loss framework typical in PPO methodologies33:
where index \(i\) represents the chosen action, and \(k\) is the worker model that updates the central model based on its localized experiences. The parameter \(\epsilon\), a crucial PPO hyperparameter, is called clipping sensitivity. It regulates the permissible deviation between the policies of the worker and the main model before any gradient clipping occurs. Further, we introduce an entropy bonus to enhance the standard policy and entropy loss equation typically employed in PPO scenarios, aiming to enrich the model’s decision-making process33.
In this configuration, incorporating the minimum term inhibits direct factorization for obtaining the scope factor. By setting \(\alpha \,=\,0\), we customize the SLF for optimal functionality within PPO frameworks33:
When the ratio \(\frac{{p}_{i}}{{p}_{i}^{k}}{A}^{k}\) achieves a minimum value, we adjust the equation to highlight the scaling factor tailored for PPO applications33.
In PPO applications, the scope scaling factor represented by (\(A-{\rm{\alpha }}{p}_{i}^{k}\log \left({p}_{i}\right)\) significantly influences gradient adjustments based on the entropy differences between the main model’s and the worker model’s policies. This scaling modifies gradients in several critical ways33:
-
If the main model shows high certainty (\({p}_{i}\)) and \(A\) is positive, this configuration dampens the gradient to prevent excessive exploitation.
-
When the worker model’s certainty (\({p}_{i}^{k}\)) is less than the main model’s (\({p}_{i}\)), and \(A\) is positive, the scaling moderates the gradient to reduce disparities between the policies of the two models.
-
Conversely, if the worker model’s certainty (\({p}_{i}^{k}\)) surpasses that of the main model and \(A\) is negative, it similarly adjusts the gradient to diminish the policy divergence.
Further, we modify the focal loss framework to align seamlessly with the PPO strategy as outlined33:
Algorithm 1
Active Learning with deep Q-network.
Input: \({X}_{L}\): Set of labeled images \(\{({x}_{{L}_{1}},\,{y}_{{L}_{1}}),\,({x}_{{L}_{2}},\,{y}_{{L}_{2}}),\,\ldots,\,({x}_{{L}_{{N}_{L}}},\,{y}_{{L}_{{N}_{L}}})\}\), \({X}_{U}\): Set of unlabeled images \(\{({x}_{{U}_{1}},\,{y}_{{U}_{1}}),\,({x}_{{U}_{2}},\,{y}_{{U}_{2}}),\,\ldots ,\,({x}_{{U}_{{N}_{U}}},\,{y}_{{U}_{{N}_{U}}})\}\)
Initialize the DQN with random parameters.
Set the limit for the number of annotations permitted
Train the initial classification model on the labeled data set \({X}_{L}\)
Loop until the stopping criterion is met:
Randomize the order of images in \({X}_{U}\) for unbiased processing.
Iterate over each image \({x}_{{U}_{i}}\) in \({X}_{U}\):
Extract the feature vector \({f}_{{U}_{i}}\) using the CNN applied to \({x}_{{U}_{i}}\)
Pass \({f}_{{U}_{i}}\) through FC layers to obtain the output \(f({x}_{{U}_{i}})\)
Combine the feature vector and the output to form the state \({s}_{i}\) = \(({f}_{{U}_{i}},f({x}_{{U}_{i}}))\)
Use the DQN to determine the action \({a}_{{\rm{i}}}\) (whether to label the image or not) based on the state \({s}_{i}\)
If the action \({a}_{{\rm{i}}}\) is to label the image:
Manually label the image \({x}_{{U}_{i}}\) to obtain \({y}_{{U}_{i}}\)
Add the newly labeled image (\({x}_{{U}_{i}}\), \({y}_{{U}_{{\rm{i}}}}\)) to the dataset \({X}_{L}\)
Assign a predefined negative reward \({r}_{i}\) (e.g., −0.2) for using the annotation
Retrain the classifier with the updated dataset \({X}_{L}\)
Otherwise:
Calculate the reward \({r}_{i}\) using a predefined equation (e.g., Eq. 3)
Update the policy of the DQN to optimize the objective, adjusting the parameters θ using:
\(\theta \leftarrow \,{\theta }_{{old}}\,+{learning\; rate}\times {\nabla }_{\theta }{Focal\; loss}\left({PPO}\right)\)
Terminate the loop if all images in \({X}_{U}\) are exhausted or the annotation limit is reached
Conclude the algorithm when the termination conditions are satisfied
Algorithm 1 outlines the pseudocode for our proposed model, which begins with an initial training phase for CNNs using a small subset of labeled data, approximately 10%. This stage provides the model with a preliminary understanding of the data characteristics. Following this, the deep Q-network (DQN) is initialized with arbitrary weights, and a cap is set on the permissible annotations within the budget. As the process unfolds, the unlabeled dataset undergoes random shuffling to ensure varied sample selection. CNNs derive feature vectors for each shuffled image, and FC layers output the present state. Utilizing this state, the DQN determines whether the image warrants labeling. If chosen for labeling, the image is annotated and added to the labeled data pool, \({X}_{L}\), incurring a penalty on the reward function to account for the cost of using annotation resources. Alternatively, if no labeling is decided, the reward is computed based on other predefined criteria. Concurrently with these decisions, the classifier is regularly retrained with newly annotated data to refine its accuracy. Meanwhile, the DQN revises its strategy, aiming to enhance the selection of future data, driven by the rewards obtained and a deepening insight into the data’s characteristics. This update cycle continues until all unlabeled images are processed or the annotation budget is exhausted. This methodology ensures that the model dynamically adjusts to new information, thereby boosting its predictive accuracy and optimizing the utilization of available labeling resources.
Hyperparameter optimization
Adjusting hyperparameters is crucial in deep learning and DRL, as it significantly impacts model training performance, efficiency, and effectiveness. Hyperparameters, such as learning rate, batch size, and network architecture, are pivotal in determining how well a model can learn from its environment and generalize to new, unseen data. In DRL, the correct tuning of hyperparameters can prevent the model from getting stuck in local minima or failing to converge. It also ensures that the agent explores the environment thoroughly while learning to make optimal decisions based on the rewards it receives. Optimal hyperparameter settings can lead to faster convergence, more stable learning, and improved overall performance, making tuning them a critical aspect of developing robust deep learning and DRL models43.
Table 1 provides a detailed list of the hyperparameters used in our research. The range of values for each was chosen based on previous studies in AD. These hyperparameters were then tuned using the DE algorithm to identify values that maximize model accuracy. We applied the optimization process simultaneously to both the Open Access Series of Imaging Studies (OASIS) and the Alzheimer’s Disease Neuroimaging Initiative (ADNI) datasets. Remarkably, the DE algorithm identified the same set of optimal hyperparameter values for both datasets, as shown in the third column of Table 1. The identical results across these two datasets demonstrate that the selected hyperparameters are robust and generalizable. This also confirms that our model performs reliably on different types of data.
Random Key
This paper adopts the Random Key approach44 for hyperparameter optimization (HO) in the proposed model due to its versatility and effectiveness in handling a diverse range of parameter types, which is crucial for complex models. The Random Key method offers a robust solution by encoding hyperparameters into a continuous numeric range, thereby simplifying the optimization process across both discrete and continuous spaces. This encoding enables the integration with various optimization algorithms, including genetic algorithms and swarm intelligence, enhancing the adaptability of the model’s hyperparameter tuning phase. The Random Key approach also facilitates a straightforward transition from the encoded numeric space to the actual hyperparameters, ensuring that the optimization process remains efficient and accurate. This method is particularly beneficial for complex models with many hyperparameters, where traditional grid or manual search methods would be computationally expensive and time-consuming. Using this approach, the model can achieve optimal performance by systematically exploring and exploiting the hyperparameter space with increased precision and less computational overhead.
The Random Key method employs an array of \(T\) numeric vectors, each with D dimensions labeled as \({p}_{1},\,{p}_{2},\,...,\,{p}_{T}\). These vectors collectively constitute a pool, where each vector symbolizes a potential solution associated with model hyperparameters through a mapping function known as the random key. For refining \(C\) hyperparameters (as outlined in Table 1), each hyperparameter from 1 to \(C\) is represented by \({D}_{c}\) slots in the vector. For continuous hyperparameters, \({D}_{c}\) is designated as one, culminating in a total dimension \(D\) calculated as \(D=\mathop{\sum }\nolimits_{c=1}^{C}{D}_{c}\). Each vector, \({p}_{i}\), is partitioned into \(C\) segments containing \({D}_{c}\) slots that propose potential values for a hyperparameter. For continuous hyperparameters, these values are generally normalized to align within the permissible parameter range. In contrast, for categorical hyperparameters, the Random Key approach converts the numeric segment \({D}_{c}\) of vector \({p}_{i}\) into a corresponding categorical array, \({{MAP}}_{c}\), which enumerates the options for the \({c}^{{th}}\) hyperparameter. This conversion is accomplished by sorting the entries within the \({D}_{c}\) segment and selecting the highest-ranked element’s position as an index into the \({{MAP}}_{c}\) array to determine the appropriate categorical value. This ensures that the results of evolutionary operations such as mutation, crossover, and selection, when applied to the numeric vectors \({p}_{i}\), are consistently translated back into configurations that support both categorical and continuous hyperparameter values. Figure 2 demonstrates this method with a \({D}_{c}\) of 5. Essentially, a random key is a vector of real numbers that, once ordered, plays a crucial role in aligning with a predefined array of choices. This setup enables more significant features to emerge within the key, enhancing the model’s ability to efficiently assess and prioritize feature importance.
Differential evolution
We enhance the Random Key encoding strategy by integrating the DE algorithm, a population-based optimization method suited for continuous, non-differentiable, and high-dimensional search spaces. One of the key advantages of DE lies in its superior performance compared to traditional hyperparameter tuning methods. For instance, grid search evaluates all predefined parameter combinations, which becomes computationally expensive as dimensionality increases. In contrast, DE performs fewer evaluations and explores the parameter space more flexibly using vector differentials. Moreover, DE demonstrates better scalability and robustness than Bayesian optimization (BO), which relies on surrogate models and acquisition functions. This advantage is especially relevant in noisy or high-dimensional landscapes, which are often encountered in DRL. BO also has higher computational costs due to repeated model fitting, while DE operates directly on the objective function and avoids such overhead. DE also performs well compared to hybrid optimization methods that combine global and local search strategies. Although hybrid methods can be effective, they often involve complex design and coordination. In contrast, DE maintains a simple yet efficient structure, utilizing mutation, crossover, and selection to achieve a strong balance between exploration and exploitation without requiring extensive parameter tuning.
Additionally, DE demonstrates advantages over other evolutionary algorithms. GAs use crossover and mutation operations but often converge prematurely. In contrast, DE guides the search using differences between randomly selected individuals. This differential-based mutation strategy encourages both exploration and diversity while reducing sensitivity to the initial parameter settings. Particle swarm optimization often converges quickly but may struggle to effectively explore complex search spaces. DE offers a better balance between global and local search, which lowers the chance of stagnation.
DE is simple and requires minimal internal hyperparameter tuning. These characteristics make it highly adaptable to problems of different sizes. DE can be combined with Random Key encoding and k-means-based candidate selection to improve performance. In this setup, DE directs mutations toward clusters with lower objective values, which helps it converge faster to optimal hyperparameters. This synergy leads to more robust hyperparameter tuning, improved convergence stability, and enhanced model performance.
The DE algorithm functions in three main stages: mutation, crossover, and selection, starting with mutation. This phase is essential for introducing new genetic diversity into the solution pool. DE modifies existing solutions by integrating elements from different population members using specific mathematical operations. Commonly, this involves modifying a base solution by adding the scaled difference between two additional solutions, thus generating a new variant with potentially improved efficacy. These mutations are crucial for maintaining diversity in the solution pool and discovering novel, superior solutions that can improve overall solution quality and prevent the algorithm from converging prematurely on suboptimal outcomes. The effectiveness of the DE algorithm largely hinges on the mutation phase’s capacity to produce a varied and high-quality set of solutions, driving the algorithm toward optimal results.
Within the DE framework, the mutation stage generates a new vector through the following procedure36:
Here, \({\vec{x}}_{{r}_{1},g}\), \({\vec{x}}_{{r}_{2},g}\), and \({\vec{x}}_{{r}_{3}}\) represent three distinct solutions randomly selected from the current population. \(F\) is a scaling factor that adjusts the influence of the differences between two solutions on the third. After mutation, the crossover phase commences, combining elements of this newly mutated vector with the existing target vector. This combination typically uses a binomial crossover method, which systematically integrates characteristics from the mutated vector into the target vector, thereby introducing new genetic diversity into the solution pool and enhancing the potential for evolutionary advancement:
In this scenario, \({CR}\) denotes the crossover rate, and \({j}_{{rand}}\) represents an integer randomly chosen from the set \(\{1,\,2,\,...,{D}\}\), where \(D\) is the dimension of the solution space. Following this, the selection phase occurs, where the original target vector and the newly formed trial vector from the crossover are evaluated. The algorithm selects the superior solution for advancement in the evolutionary process. This selection step is crucial as it promotes the continuous enhancement of solutions within the population, ensuring that only the most effective solutions are retained.
The enhanced DE variant introduces an innovative mutation mechanism informed by recent advancements in the field45. Initially, k-means clustering is applied to the population to delineate distinct groups within the search space. This segmentation divides the population into several clusters, with the number of clusters, \(k\), randomly set to range between \([2,\,\sqrt{N}]\). The focus is then placed on the cluster exhibiting the smallest average objective function value, which is identified as the primary target for in-depth analysis.
We present a new mutation operator influenced by clustering, which is formulated as follows:
In this setup, \({\vec{x}}_{{r}_{1},g}\) and \({\vec{x}}_{{r}_{2},g}\) denote two randomly selected candidate solutions from the group, while \({\overrightarrow{{win}}}_{g}\) signifies the optimal solution within a particularly promising cluster. It is important to note that \({\overrightarrow{{win}}}_{g}\) may not consistently represent the best solution for the entire group. This cluster-focused mutation approach is repeatedly executed over \(M\) iterations. Subsequently, the population progresses through several stages as outlined by the generic population-based algorithm (GPBA)46:
-
Selection: \(k\) solutions are randomly chosen to serve as initial centroids for the k-means clustering process.
-
Generation: \(M\) new candidate solutions are continuously generated through mutation, emphasizing the ongoing evolution of the algorithm, forming the group \({v}^{{clu}}\).
-
Replacement: \(M\) candidates are randomly selected from the current population to compose a new set, referred to as \(B\).
-
Update: The top \(M\)-performing candidates from the merged pools of \({v}^{{clu}}\) and \(B\) are selected to form the new set \(B^{\prime}\). The updated population is then reassembled by combining \(P-B\) with \({B}^{{\prime} }\).
Results
The section commences by scrutinizing the dataset, detailing its characteristics and relevance to our study. Subsequently, it delves into the metrics, explicating the principal standards and assessment criteria employed to evaluate the performance of our models. The results are then presented, highlighting the crucial discoveries from our experiments and analyses and discussing their implications for our research goals.
Datasets
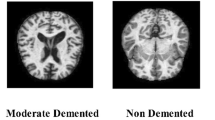
In this study, we employ the OASIS47 and ADNI datasets48 to assess the effectiveness of the proposed model. Figure 3 displays examples of both normal and Alzheimer’s cases from each dataset. An overview of these datasets is provided as follows.
-
The OASIS dataset is a valuable resource for neuroscientific research, offering researchers high-quality brain imaging data at no cost. It collates a wide array of neuroimaging data to support developments in scientific and clinical brain studies. Specifically, it facilitates neurological, clinical, and cognitive investigations into normal aging processes and cognitive disorders, encompassing a range of neurological conditions and genetic backgrounds. Within our study, the dataset comprises 436 brain scans, consisting of 98 AD-positive and 338 normal MRI images, available in coronal, sagittal, and axial planes. The images are divided into four categories: Mildly Demented, Very Mildly Demented, Moderately Demented, and Cognitively Normal. Given the unequal distribution of images across these categories, the study simplifies the classification into a binary system. It groups the Mildly Demented, Very Mildly Demented, and Moderately Demented into a single AD-positive category (labeled as 1). At the same time, the Cognitively Normal images are categorized as AD-negative (labeled as 0). This binary classification approach streamlines the learning process by consolidating the AD-positive categories, facilitating more effective training on the diverse conditions represented within the AD-positive group.
-
The ADNI primarily examines whether serial MRI, PET scans, and various other biological markers, along with clinical and neuropsychological assessments, can effectively track the progression of mild cognitive impairment (MCI) and early AD. The findings from the ADNI are vital for propelling forward clinical research focused on the early diagnosis, prevention, and treatment of Alzheimer’s disease. This initiative is crucial in pinpointing biomarkers essential for accurate diagnosis and ongoing AD monitoring, made accessible through its open datasets. ADNI plays a key role by supplying extensive long-term MRI and PET scans of elderly individuals diagnosed with AD, MCI, and similar conditions. For our analysis, we extracted 321 MRI images labeled as Cognitively Normal (CN) and 136 MRI images identified as AD from the ADNI database. These images are divided for binary classification, where CN images are labeled as AD-negative (0) and AD images as AD-positive (1), amounting to 457 images utilized in our study.
Metrics
We employ a robust set of metrics, including accuracy, F-measure, geometric mean (G-mean), and Area under the curve (AUC)49, to evaluate the model, each chosen for its ability to provide a comprehensive assessment of classifier performance from different perspectives. Accuracy is fundamental, as it measures the overall correctness of the model across all classifications, providing a straightforward indication of performance. The F-measure is beneficial in imbalanced datasets as it balances precision and recall, offering insight into how effectively the model discriminates between classes. The g-mean measures the balance between the performance of the majority and minority classes, ensuring that improvements in model performance do not come at the expense of minority class recognition. Lastly, the AUC of the receiver operating characteristic is a robust metric for evaluating the probability that a classifier will rank a randomly chosen positive instance higher than a randomly chosen negative instance, regardless of the class distribution, providing a performance measure that is invariant to threshold shifts and class imbalances. Together, these metrics ensure a rounded assessment that captures the model’s strengths and potential weaknesses across varied data scenarios.
The Accuracy, F-measure, G-means, and AUC metrics are defined as follows:
where
Main evaluation
The study was conducted on a 64-bit Windows 10 operating system using Python 3.8. The deep learning models were implemented with TensorFlow 2.4 and Keras 2.4.3 libraries to facilitate the construction and training of the CNNs. The reinforcement learning aspects were managed using the stable-baselines3 library, which provides an efficient and stable framework for DRL tasks. For the DE algorithm, we utilized the SciPy library, version 1.6, which offers robust tools for optimization, including our custom implementations for the mutation mechanisms and integration with k-means clustering. Hardware-wise, the computations were performed on an Intel Core i9 processor with 32 gigabytes (GB) of random-access memory (RAM) and an NVIDIA GeForce RTX 3080 graphics processing unit (GPU), significantly accelerating the training and evaluation processes. This setup ensured that the resource-intensive tasks of training deep learning models and running complex simulations could be completed efficiently. Our choice of hardware and software was guided by the need for high computational power and precision, which are essential for handling large datasets like OASIS and ADNI, as well as performing advanced image processing and machine learning tasks. All experiments, including the proposed model and baseline methods, were conducted under identical hardware and software settings to ensure fair and consistent comparisons.
To assess the performance and reliability of our models, we employed a 5-fold stratified cross-validation across all results. This method is particularly effective in ensuring that each fold of the dataset is a good representative of the whole, maintaining the same proportion of class labels across each subset. This is crucial in medical imaging studies like ours, where the class distribution can be significantly imbalanced. In our setting, the labeled and unlabeled data are derived from the same dataset, ensuring consistency in the evaluation process. Stratified cross-validation helps to prevent overfitting and ensures that the evaluation metrics are reliable and consistent across different subsets of the data. By using this approach, we can confidently generalize the performance of our model, as it mitigates the risk of biases that could arise from random sampling, especially when dealing with smaller or unevenly distributed datasets. Each performance metric is reported in the format of mean ± standard deviation, derived from 5-fold stratified cross-validation.
During the assessment phase, our model was rigorously compared against eight other cutting-edge models: HBOA-MLP19, CNN20, VGG1621, CLADSI22, DQN29, 4D-AlzNet50, XAI51, and RESNET50 KNN52. We also performed ablation studies to evaluate the impact of incorporating active learning (AL), SLF, and HO into our model. The results of these thorough evaluations across the OASIS and ADNI datasets are presented in Tables 2 and 3.
On the OASIS dataset, RESNET50 KNN achieved the highest performance among eight advanced models, with 84.179% accuracy and an 87.068% F-measure. Its success is attributed to RESNET50’s strong feature extraction capabilities and KNN’s effective classification, although its high computational cost limits its use in real-time applications. The proposed model, which incorporates AL, SLF, and HO, outperformed RESNET50 KNN. It improved accuracy by 5.5%, F-measure by 5.7%, and AUC by 12.6%. This demonstrates the strength of combining deep learning with targeted optimization. Removing any of the components (AL, SLF, or HO) led to decreased performance. For example, accuracy dropped from 88.806% without HO to 87.739%. This highlights HO’s crucial role in adapting to data complexity. Similarly, SLF and AL proved critical, with SLF refining feature focus and AL enabling efficient learning from key samples. These findings underscore the need for integrated optimization strategies to maximize model performance.
In the ADNI dataset analysis, the proposed model consistently outperformed other models. RESNET50 KNN performed strongly, achieving 87.438% accuracy and a 91.602% F-measure, reflecting its robustness due to deep architecture and KNN’s strength in high-dimensional spaces. However, the proposed model surpassed these results, improving accuracy by 3.6%, F-measure by 2.3%, and AUC by 7.9%. This improvement highlights its superior generalization ability across diverse medical images. Comparing the proposed model with its derivatives, the complete model outperformed versions without specific enhancements. For instance, without HO, accuracy dropped to 88.071%, while the full model achieved 90.623%, a 2.9% gain. Similarly, removing SLF and AL also reduced performance, confirming the critical role of each component. These enhancements are significant for the ADNI dataset, where noise and imaging variability can challenge model accuracy. The proposed model’s integration of advanced techniques ensures strong adaptability and performance across complex medical imaging scenarios.
The proposed model exhibits lower standard deviations across all evaluation metrics compared to methods such as HBOA-MLP, VGG16, and CLADSI on both the OASIS and ADNI datasets. This low variability reflects the consistent performance of the model across different cross-validation folds. Such stability indicates strong generalization ability and reduced sensitivity to data partitioning. These low values confirm that the model performs reliably regardless of how the training data is split, which supports the robustness of the proposed architecture under different experimental conditions.
To validate the superiority of the proposed model, paired t-tests were performed across accuracy, F-measure, G-means, and AUC metrics. On the OASIS dataset, the proposed model significantly outperformed others, with p-values below 0.01 for comparisons against RESNET50 KNN. Confidence intervals did not overlap, reinforcing the robustness of the results. Ablation studies demonstrated that removing components such as AL, SLF, or HO resulted in statistically significant performance drops. These results confirm the necessity of full integration. Similarly, for the ADNI dataset, the proposed model consistently achieved statistically significant improvements, with p-values of less than 0.01 when compared to RESNET50 KNN. Removing any component caused performance degradation, with p-values of less than 0.05. These findings, supported by low p-values and non-overlapping confidence intervals, affirm the model’s superiority. They also highlight the critical contributions of AL, SLF, and HO to achieving optimal results across different datasets and evaluation metrics.
Tables 4 and 5 present an analysis of the computational efficiency of the evaluated models, including runtime and GPU usage, during the training and testing phases on the OASIS and ADNI datasets. During training on the OASIS dataset, the DQN model achieved the shortest runtime, completing in 2850 s. HBOA-MLP closely followed it at 2863 s and the proposed model at 3056 s. The proposed model outperformed more resource-heavy models like VGG16 and RESNET50 KNN. In GPU usage, the proposed model consumed 22.1 GB, higher than XAI (17.6 GB) but lower than RESNET50 KNN (24.9 GB). On the ADNI dataset, the proposed model again demonstrated strong efficiency, with a runtime of 2930 s. It was just behind CLADSI and significantly faster than RESNET50 KNN. GPU usage was 23.4 GB, which is well-balanced compared to the highest-consuming VGG16 model.
During testing, the proposed model achieved a runtime of 12.5 ms on the OASIS dataset and 13.5 ms on the ADNI dataset, placing it among the more efficient models. The model used slightly more GPU memory (12.5 GB and 9.6 GB) than lightweight models such as CNN and XAI. However, it required substantially less memory than deeper architectures, such as VGG16 and ResNet50 KNN. This efficiency during inference and training can be attributed to the streamlined architecture of the proposed model, which integrates CNNs with FC layers. Unlike resource-intensive models such as VGG16 and RESNET50 KNN, which employ very deep or transfer learning-based architectures and exhibit higher runtime and GPU consumption, the proposed model achieves competitive accuracy with a significantly lower computational burden. This compact design reduces overhead during inference and training while maintaining strong performance.
Figure 4 illustrates the training and validation loss trajectories of the proposed model over 250 epochs on the OASIS and ADNI datasets. For the OASIS dataset, both losses decline sharply during the initial epochs and then stabilize. The validation loss remains close to the training loss, suggesting effective generalization and minimal risk of overfitting. Similarly, the ADNI dataset shows a rapid early decrease in losses, with the validation curve closely tracking the training curve. The consistent closeness and convergence of the training and validation curves across epochs indicate that the model learns generalizable patterns rather than memorizing specific data points. Toward the later epochs, both losses remain relatively stable, reinforcing the model’s strong generalization capability. Overall, the parallel decrease in training and validation losses across both datasets highlights the proposed model’s robustness and suitability for real-world deployment, where reliable performance on new, unseen data, such as inpatient diagnosis, is critical.
Figure 5 compares the accuracy trajectories of the proposed model with and without active learning across the OASIS and ADNI datasets. On the OASIS dataset, the model with active learning demonstrated a sharp increase in accuracy from 65% to 88% as more labeled data was included. In comparison, the model without active learning only reached around 72%. This highlights the efficiency of active learning in leveraging labeled samples. A similar but more pronounced pattern emerged in the ADNI dataset. Although the active learning model started at 60%, it quickly surpassed the non-active learning model, achieving nearly 90% accuracy compared to approximately 70% without active learning. This greater gap in ADNI highlights the adaptability of active learning to complex datasets. The analysis reveals that incorporating active learning significantly enhances model performance across both datasets, enabling higher accuracy with fewer labeled examples and demonstrating its robustness in data-scarce environments.
Figure 6 illustrates the distribution of decision-making times for the proposed model on the OASIS and ADNI datasets within real-time bidding (RTB) environments. For the OASIS dataset, most decisions occurred between 60 and 80 ms, with a peak at around 70 ms, demonstrating fast and consistent performance. A slight tail beyond 80 ms suggests occasional complexity in the input data. Similarly, the ADNI dataset exhibited decision times clustering around 70 ms. However, the slightly broader spread indicates greater variability in the dataset. Despite this, the model maintained most decision times within the 60–80 ms range, confirming its robustness. Overall, the model demonstrated efficient and reliable decision-making across both datasets, making it highly suitable for RTB environments where rapid processing is crucial. Its ability to maintain consistent speed across datasets with different characteristics highlights its adaptability. This ensures that the model can meet real-time demands without compromising accuracy or reliability.
Examination of the robustness
To assess the resilience and flexibility of our proposed model against adversarial threats, we employed the Fast Gradient Sign Method (FGSM)53 to test our model on the OASIS and ADNI datasets, evaluating its performance when confronted with adversarially modified inputs. These modifications are designed to mimic potential real-world disturbances, making such tests crucial for verifying the dependability of neural network models under adversarial conditions.
The results are shown in Tables 6 and 7 for the OASIS and ADNI datasets. FGSM-based adversarial testing on the OASIS dataset revealed that all models experienced performance drops, underscoring the challenges posed by adversarial attacks. Baseline models, such as HBOA-MLP and CNN, were less resilient, while VGG16 and CLADSI showed moderate robustness. Advanced models, such as XAI and ResNet50 KNN, performed better, achieving an accuracy of 75.549%. Notably, the proposed model achieved an accuracy of 83.838%, nearly 8% higher than that of RESNET50 KNN. This improvement demonstrates strong adversarial defense, supported by enhanced feature extraction and learning strategies. Similar trends were observed in the ADNI dataset, where simpler models struggled, while complex models, such as XAI and RESNET50 KNN, maintained accuracy above 80%. Again, the proposed model excelled, achieving 86.236% accuracy, about 5% higher than RESNET50 KNN, showing superior resilience. These results across both datasets confirm that the proposed model performs well under adversarial conditions. This makes it a strong candidate for secure and reliable deployment in critical real-world applications such as medical imaging analysis.
Impact of the RL method
Figure 7 illustrates the benefit of incorporating the SLF into the proposed model over 250 epochs, specifically highlighting the model’s performance with the OASIS and ADNI datasets. For the OASIS dataset, the graph shows that using the SLF provides a clear advantage. While both models start with high loss values, the model with scope loss rapidly and steadily declines, stabilizing around mid-epochs, whereas the model without it shows ongoing fluctuations. These fluctuations suggest difficulty in adapting without scope loss. The smoother decline with scope loss highlights its role in leveraging known data and integrating new insights. This process leads to improved model stability and learning efficiency. Similar trends are observed in the ADNI dataset, albeit with slightly higher initial loss due to its complexity. Again, the model with scope loss exhibits steadier, more robust loss reduction, while the model without it shows greater volatility and sustained higher loss. These observations affirm that the SLF significantly enhances model stability, learning efficiency, and adaptability across complex datasets. They confirm its pivotal role in optimizing performance in reinforcement learning applications.
Figure 8 presents the cumulative reward trajectories of the reinforcement learning agent over 250 episodes on the OASIS and ADNI datasets. For the OASIS dataset, the agent shows a consistent upward trend with subtle fluctuations, indicating steady learning progress while occasionally encountering complex scenarios. The gradual slope suggests incremental learning. The small peaks and troughs reflect temporary challenges that the agent eventually overcomes. In contrast, the ADNI dataset exhibits a steeper and smoother cumulative rewards trajectory. This steeper ascent suggests that the agent adapts and optimizes strategies more effectively in the ADNI environment. This may be due to lower data variability or better alignment with the agent’s learning mechanisms. The smoother curve also indicates a more stable learning process. Overall, the analysis highlights the agent’s ability to learn steadily in complex environments, such as OASIS, and to achieve faster, more stable learning on structured datasets, like ADNI, showcasing its adaptability and robust performance.
Analysis of the proposed DE algorithm
In the next experiment, we present a comparative analysis of the proposed DE algorithm with several established metaheuristic optimization techniques, focusing specifically on their performance in hyperparameter tuning across the OASIS and ADNI datasets. The competing techniques include the firefly algorithm (FA)54, salp swarm algorithm (SSA)55, bat algorithm (BA)56, Cuckoo optimization algorithm (COA)57, human mental search (HMS)58, artificial bee colony (ABC)59, and original DE. Tables 8 and 9 present the outcomes corresponding to the OASIS and ADNI datasets.
The comparative evaluation using the OASIS dataset highlights the superiority of the proposed DE algorithm over other metaheuristic methods. FA and SSA achieved moderate accuracies (77.479% and 79.171%), while BA and COA performed slightly better (80.555% and 81.543%). HMS and ABC improved performance, reaching 83.088% and 83.827% accuracy, respectively. However, the proposed DE algorithm achieved the highest accuracy of 88.806%, demonstrating superior optimization through effective navigation of the search space. Similar trends were observed on the ADNI dataset, with generally higher performance across all algorithms. ABC performed strongly with 84.768% accuracy, but the proposed DE algorithm outperformed all others with 90.623%. These results confirm the DE algorithm’s consistent advantage in handling complex optimization challenges. This advantage is attributed to its robust exploration-exploitation balance and adaptive parameter strategies. This finding reinforces the broader applicability of the DE algorithm for optimizing hyperparameters in data-driven applications. It is beneficial when data inconsistency and sensitivity are critical concerns.
Figure 9 visualizes the loss reduction over 250 iterations using the DE algorithm on the OASIS and ADNI datasets. For the OASIS dataset, the loss declines moderately and steadily, starting from a higher initial value. This suggests that optimization challenges may arise from complex data patterns or less-than-ideal initial parameters. Although the DE algorithm gradually reduces the loss, the slower rate of decline and slight fluctuations suggest that the algorithm is continuously adapting to varying data complexities. In contrast, the ADNI dataset exhibits a steeper and more consistent loss reduction from the outset, suggesting a better alignment between the data characteristics and the DE algorithm’s optimization strategy. The quicker decline and fewer fluctuations suggest that ADNI data is potentially less complex or more uniform, enabling faster learning and adaptation. Overall, the DE algorithm optimizes both datasets, achieving more rapid and stable improvements in ADNI, which highlights the influence of dataset complexity on optimization dynamics.
Figure 10 shows the impact of six key continuous hyperparameters on model accuracy, providing insight into their optimal configurations. Each sub-plot shows how model accuracy changes as the value of a single hyperparameter varies, while all other hyperparameters are held constant at their optimal values. The curves reveal clear peaks, indicating the best value for each hyperparameter. For example, accuracy is highest when the batch size is 71, the learning rate is 0.002, the number of epochs is 278, the number of MLP layers is 4, the number of CNN stacks is 3, and the \(\gamma\) value is 0.2. As seen in the plots, deviating from these values in either direction lowers performance, highlighting the importance of careful tuning. These values correspond to the optimal configurations identified by the DE algorithm, as presented in Table 1. This confirms the ability of DE to efficiently explore the hyperparameter space and choose settings that improve both accuracy and training stability.
Discussion
This study presents a new framework for the early detection of AD using DRL, SLF, and DE. A key advantage of the model is its high diagnostic accuracy combined with reduced reliance on large annotated datasets. To address this, we use DRL-based active learning, which adapts dynamically, unlike many static models. The SLF component enhances sample selection by striking a balance between data exploitation and exploration. The DE algorithm addresses hyperparameter sensitivity, a common limitation in DRL-based models. To our knowledge, this study is the first to integrate DRL-based active learning with SLF and DE into a unified framework for AD detection. This integration represents a novel contribution to the field of early AD diagnosis. The proposed model achieved F-measures of 92.044% and 93.685% on the OASIS and ADNI datasets, respectively, outperforming baseline models. These components make the framework especially useful in clinical settings where annotated data are limited and early diagnosis is essential. Beyond AD, the framework may also generalize to other neurodegenerative diseases and imaging tasks with similar constraints.
The comparative analysis in Tables 2 and 3 illustrates the model’s superior performance against a suite of state-of-the-art models across the OASIS and ADNI datasets. For better clarity, Table 10 summarizes the results obtained by the proposed model compared to the best existing method (RESNET50 KNN52), along with the performance improvement achieved by the proposed model. Notably, the model achieves remarkable improvements in accuracy, F-measure, G-means, and AUC metrics, significantly outperforming other established methods. These findings are crucial when placed within the broader literature on AD detection. Recent studies have emphasized the need for highly accurate and efficient models due to the challenging nature of AD pathology. The integration of DRL and SLF addresses critical issues in model training dynamics by optimizing the balance between the exploration of new data and the exploitation of known data, which is often a limitation in other models. Furthermore, using a DE algorithm for hyperparameter tuning is particularly effective, as evidenced by the performance metrics, and represents an advancement over more traditional optimization methods often used in medical imaging analysis.
Integrating active learning with RL is designed to overcome the inefficiencies of traditional active learning methods, which often rely on a heuristic selection unrelated to the model’s learning progress. By adopting RL, the model dynamically identifies the most informative unlabeled samples that, once labeled, are expected to yield the highest benefit in terms of learning. This strategic selection process is driven by RL’s ability to make sequential decisions, thus optimizing the learning trajectory by continuously adapting to the newly acquired data. This approach ensures that each decision to label a new data point is based on its potential to enhance the overall model performance. It efficiently uses limited resources by focusing on the most significant incremental value samples.
The SLF function is a pivotal enhancement within the RL framework aimed at balancing the exploration of new data against the exploitation of already known data. This dual-objective function helps mitigate the common RL dilemma between exploring new strategies and refining known ones. By effectively integrating SLF, the model can maintain a delicate balance, prioritizing exploration to discover potentially superior strategies or focusing on exploitation to maximize immediate performance from current knowledge. The SLF accelerates the learning process by minimizing excessive exploration and stabilizes the learning curve, thereby enhancing the robustness and reliability of the model’s predictions.
The enhanced DE algorithm employed in this model introduces a novel mutation mechanism integrated with k-means clustering, specifically designed to optimize the hyperparameters with utmost precision. This adaptation enables a more precise and efficient exploration of the hyperparameter space, ensuring that model configurations are continually optimized to address the complex and variable nature of medical imaging data. The use of k-means clustering helps identify and focus on the most promising regions of the hyperparameter space, thereby reducing computational overhead and accelerating convergence towards the optimal set of parameters. This strategic improvement in the DE algorithm significantly contributes to the overall precision and efficiency of the model, ensuring optimal performance even with dynamically changing data conditions.
While the proposed model achieves strong diagnostic performance, it also involves some trade-offs that need to be considered. Using multiple CNNs, RL, SLF, and DE improves accuracy and flexibility, especially when only a small number of labeled samples are available. However, this improved performance comes at the cost of increased computational complexity, particularly during the training stage. The model is more efficient than many conventional deep learning methods; however, its high computational demand may compromise its reliability in real-time or low-resource environments. These trade-offs underscore the need to further refine the framework to achieve a more balanced approach between accuracy, robustness, and efficiency for clinical applications.
An alternative solution to the problem of limited access to AD training image datasets is transfer learning using CNNs pre-trained on large open-source annotated visual databases60. Several transfer learning models for MRI-based AD detection and/or grading50,51,52,61,62,63 have been published. While efficient, transfer learning faces the domain adaptation problem, where a model trained on a source dataset may not perform well on a target dataset due to differing dataset characteristics. Active learning tackles this by selectively querying for labels from the most informative data points in the target domain, enabling more accurate adjustments to new data features. Another challenge is catastrophic forgetting, where a model must retain knowledge acquired from the source dataset when introduced to new data. Active learning addresses this by balancing training inputs from old and new data, thereby preserving essential information from the source while adapting to the target dataset. Lastly, transfer learning may yield biased predictions if the source and target datasets are not closely aligned. In contrast, active learning promotes model robustness by iteratively soliciting diverse and representative samples from within the target dataset, thereby enhancing model generalizability across various data scenarios.
The versatility of the proposed model makes it highly applicable not only to AD but also to other neurodegenerative disorders and broader medical imaging tasks. For example, conditions such as Parkinson’s disease, Huntington’s disease, and multiple sclerosis (MS) rely heavily on early diagnosis through imaging techniques like MRI. These diseases involve gradual structural changes in the brain that require sensitive and efficient detection methods. The model can learn effectively even with limited labeled data. It also handles complex image patterns well, making it suitable for these applications. The model uses an adaptive learning framework that can be fine-tuned for different diseases. This is done by incorporating disease-specific data characteristics, which helps maintain reliable performance in different clinical domains. Additionally, outside of neurodegenerative disorders, the model could be extended to detect early-stage cancers in oncology, where labeled datasets are often scarce. This adaptability highlights the model’s strong potential for generalization across a wide range of clinical imaging tasks.
The list of assumptions considered in the paper is as follows:
-
It is assumed that the MRI images in the OASIS and ADNI datasets are of sufficient quality and consistency to support accurate feature extraction and classification.
-
It is assumed that a limited number of labeled samples are sufficient to train an effective classifier when guided by the proposed active learning and reinforcement learning framework.
-
It is assumed that the selected features extracted by the CNNs capture the relevant structural changes associated with AD progression.
-
It is assumed that the distribution of labeled and unlabeled samples is representative of real-world clinical variability and does not introduce significant bias.
-
It is assumed that the learned model can generalize across different patients and scanning conditions within the OASIS and ADNI datasets.
-
It is assumed that the integration of scope loss and deep reinforcement learning effectively balances exploration and exploitation during the sample selection process.
-
It is assumed that the hyperparameter optimization process via the DE algorithm converges to globally optimal or near-optimal settings that apply to both datasets.
The limitations of the model can be summarized as follows:
-
Dependency on high-quality data: The proposed model’s effectiveness heavily relies on the availability of high-quality, well-annotated imaging data. Since the CNNs require detailed and accurate inputs to learn effectively, any inconsistencies, noise, or errors in the data can lead to significant inaccuracies in detection. This reliance on high-quality data means the model’s performance could be severely compromised in environments where data collection could be more consistent or better. To mitigate this limitation, further research could focus on developing robust preprocessing methods that enhance data quality or designing algorithms that can learn effectively from lower-quality inputs.
-
Computational resources: The proposed model is faster and more efficient than some deep architectures. However, it still utilizes multiple CNNs and advanced components, such as DRL and DE. These parts require considerable computational resources during both training and deployment. As a result, the model may not be suitable for environments with limited hardware resources, such as those found in developing countries or smaller clinics. To mitigate this limitation, future work can optimize the architecture using methods such as pruning, quantization, or knowledge distillation. These techniques lower computational requirements while maintaining performance.
-
Generalization across different imaging technologies: While the proposed model demonstrates strong potential for use in various neurodegenerative diseases and clinical imaging tasks, it has been tested only on the OASIS and ADNI datasets. These datasets use fixed imaging protocols. Other imaging types or settings might create data variations unfamiliar to the model. Although the model design is flexible, it has not yet been tested on other imaging technologies. Expanding training with more diverse datasets or utilizing domain adaptation methods would enhance the model’s robustness and make it suitable for a broader range of clinical environments.
Data availability
This study used data from the publicly available OASIS (https://www.oasis-brains.org/) and ADNI (http://adni.loni.usc.edu/) datasets.
Code availability
The source code of this paper is available at https://github.com/ZhisenHe/Alzheimer-Disease-Detection-/tree/main.
References
Xu, L., Liu, R., Qin, Y. & Wang, T. Brain metabolism in Alzheimer’s disease: biological mechanisms of exercise,. Transl. Neurodegener. 12, 33 (2023).
Ribarič, S. Detecting early cognitive decline in alzheimer’s disease with brain synaptic structural and functional evaluation, Biomedicines 11, 355 (2023).
Marvi, F., Chen, Y.-H. & Sawan, M. Alzheimer’s disease diagnosis in the preclinical stage: normal aging or dementia. IEEE Rev. Biomed. Eng. 18, 74–92 (2025).
van der Thiel, M. M., Backes, W. H., Ramakers, I. H. & Jansen, J. F. Novel developments in non-contrast enhanced MRI of the perivascular clearance system: what are the possibilities for Alzheimer’s disease research?. Neurosci. Biobehav. Rev. 144, 104999 (2023).
Hazarika, R. A. et al. An approach for classification of Alzheimer’s disease using deep neural network and brain magnetic resonance imaging (MRI). Electronics . 12, 676 (2023).
Tamburini, B. et al. Emerging roles of cells and molecules of innate immunity in Alzheimer’s disease. Int. J. Mol. Sci. 24, 11922 (2023).
Ul Amin, S., Kim, B., Jung, Y., Seo, S. & Park, S. Video anomaly detection utilizing efficient spatiotemporal feature fusion with 3D convolutions and long short‐term memory modules. Adv. Intell. Syst. 6, 2300706 (2024).
Ul Amin, S. et al. EADN: an efficient deep learning model for anomaly detection in videos. Mathematics 10, 1555 (2022).
Ul Amin, S., Kim, Y., Sami, I., Park, S. & Seo, S. An efficient attention-based strategy for anomaly detection in surveillance video. Comput. Syst. Sci. Eng. 46, 3939–3958 (2023).
Vrahatis, A. G. et al. Revolutionizing the early detection of Alzheimer’s disease through non-invasive biomarkers: the role of artificial intelligence and deep learning. Sensors 23, 4184 (2023).
Elazab, A. et al. Alzheimer’s disease diagnosis from single and multimodal data using machine and deep learning models: achievements and future directions. Expert Syst. Appl. 255, 124780, (2024).
Sudharsan, M. & Thailambal, G. Alzheimer’s disease prediction using machine learning techniques and principal component analysis (PCA). Mater. Today. Proc. 81, 182–190 (2023).
Shukla, G. P. et al. Diagnosis and detection of Alzheimer’s disease using learning algorithm,. Big Data Min. Anal. 6, 504–512 (2023).
Kaplan, E. et al. ExHiF: Alzheimer’s disease detection using exemplar histogram-based features with CT and MR images,. Med. Eng. Phys. 115, 103971 (2023).
Kumar, M. S., Azath, H., Velmurugan, A., Padmanaban, K. & Subbiah, M. Prediction of Alzheimer’s disease using hybrid machine learning technique. in AIP Conference Proceedings Vol. 2523, no. 1 (AIP Publishing, 2023).
Pasnoori, N., Flores-Garcia, T. & Barkana, B. D. Histogram-based features track Alzheimer’s progression in brain MRI. Sci. Rep. 14, 257 (2024).
Hassan, N., Musa Miah, A. S. & Shin, J. Residual-based multi-stage deep learning framework for computer-aided Alzheimer’s disease detection. J. Imaging 10, 141 (2024).
Menagadevi, M., Mangai, S., Madian, N. & Thiyagarajan, D. Automated prediction system for Alzheimer detection based on deep residual autoencoder and support vector machine. Optik 272, 170212 (2023).
Chelladurai, A., Narayan, D. L., Divakarachari, P. B. & Loganathan, U. fMRI-Based Alzheimer’s disease detection using the SAS method with multi-layer perceptron network. Brain Sci. 13, 893 (2023).
El-Assy, A., Amer, H. M., Ibrahim, H. & Mohamed, M. A novel CNN architecture for accurate early detection and classification of Alzheimer’s disease using MRI data. Sci. Rep. 14, 3463 (2024).
Arafa, D. A., Moustafa, H. E.-D., Ali, H. A., Ali-Eldin, A. M. & Saraya, S. F. A deep learning framework for early diagnosis of Alzheimer’s disease on MRI images. Multimed. Tools Appl. 83, 3767–3799 (2024).
Bringas, S., Duque, R., Lage, C. & Montaña, J. L. CLADSI: deep continual learning for Alzheimer’s disease stage identification using accelerometer data. IEEE J. Biomed. Health Inform. 28, 3401–3410 (2024).
Hasan, M. E. & Wagler, A. New convolutional neural network and graph convolutional network-based architecture for AI applications in Alzheimer’s disease and dementia-stage classification. AI 5, 342–363 (2024).
Moravvej. S. V. et al. RLMD‐PA: a reinforcement learning‐based myocarditis diagnosis combined with a population‐based algorithm for pretraining weights. Contrast Media Mol. Imaging. 2022, 8733632 (2022).
Danaei. S. et al. Myocarditis diagnosis: a method using mutual learning-based ABC and reinforcement learning. In Proc. IEEE 22nd International Symposium on Computational Intelligence and Informatics and 8th IEEE International Conference on Recent Achievements in Mechatronics, Automation, Computer Science and Robotics (CINTI-MACRo) 000265–000270 (IEEE, 2022).
Amin, S. U., Hussain, A., Kim, B. & Seo, S. Deep learning based active learning technique for data annotation and improve the overall performance of classification models. Expert Syst. Appl. 228, 120391 (2023).
Vale, I. & Barbosa, A. Active learning strategies for an effective mathematics teaching and learning. Eur. J. Sci. Math. Educ. 11, 573–588 (2023).
Liao, X., Tu, B., Li, J. & Plaza, A. Class-wise graph embedding-based active learning for hyperspectral image classification. In IEEE Transactions on Geoscience and Remote Sensing (IEEE, 2023).
Raj, A. & Mirzaei, G. Reinforcement-learning-based localization of hippocampus for Alzheimer’s disease detection,. Diagnostics 13, 3292 (2023).
Hatami, M., Yaghmaee, F. & Ebrahimpour, R. Investigating the potential of reinforcement learning and deep learning in improving Alzheimer’s disease classification. Neurocomputing 597, 128119 (2024).
Hashmi, A. & Barukab, O. Dementia classification using deep reinforcement learning for early diagnosis. Appl. Sci. 13, 1464 (2023).
Moravvej, S. V. et al. RLMD-PA: a reinforcement learning-based myocarditis diagnosis combined with a population-based algorithm for pretraining weights. Contrast Media Mol. Imaging. 2022, 8733632 (2022).
Burhani, H., Shi, X. Q., Jaegerman, J. & Balicki, D. Scope loss for imbalanced classification and RL exploration. arXiv preprint arXiv:2308.04024, (2023).
Ogunsanya, M., Isichei, J. & Desai, S. Grid search hyperparameter tuning in additive manufacturing processes. Manuf. Lett. 35, 1031–1042 (2023).
Vincent, A. M. & Jidesh, P. An improved hyperparameter optimization framework for AutoML systems using evolutionary algorithms. Sci. Rep. 13, 4737 (2023).
Moravvej, S. V., Mousavirad, S. J., Oliva, D., Schaefer, G. & Sobhaninia, Z. An improved de algorithm to optimise the learning process of a Bert-based plagiarism detection model. In Proc. IEEE Congress on Evolutionary Computation (CEC) 1–7 (IEEE, 2022).
Patel, R. A. Q. & Mihailescu, R.-C. Reducing labeling costs in Alzheimer’s disease diagnosis: a study of semi-supervised and active learning with 3D medical imaging. In Proc. International Conference on Modeling, Simulation & Intelligent Computing (MoSICom) 264–269 (IEEE, 2023).
Wang, T. & Dai, Q. A patch distribution-based active learning method for multiple instance Alzheimer’s disease diagnosis. Pattern Recognit. 150, 110341 (2024).
Hatami, M., Yaghmaee, F. & Ebrahimpour, R. Improving Alzheimer’s disease classification using novel rewards in deep reinforcement learning. Biomed. Signal Process. Control 100, 106920 (2025).
Hui, H. Y., Ran, A. R., Dai, J. J. & Cheung, C. Y. Deep reinforcement learning-based retinal imaging in Alzheimer’s disease: potential and perspectives. J. Alzheimer’s. Dis. 94, 39–50 (2023).
Dina, A. S., Siddique, A. & Manivannan, D. A deep learning approach for intrusion detection in Internet of Things using focal loss function. Internet Things 22, 100699 (2023).
Simchowitz, M. & Slivkins A. Exploration and incentives in reinforcement learning. Oper. Res. (2023).
Feurer, M. & Hutter, F. Hyperparameter optimization. Automated Machine Learning: Methods, Systems, Challenges 3–33 (Springer International Publishing, 2019).
Bean, J. C. Genetic algorithms and random keys for sequencing and optimization. ORSA J. Comput. 6, 154–160 (1994).
Mousavirad, S. J. et al. An enhanced differential evolution algorithm using a novel clustering-based mutation operator. In Proc. IEEE International Conference on Systems, Man, and Cybernetics (SMC) 176–181 (IEEE, 2021).
Deb, K. A population-based algorithm-generator for real-parameter optimization. Soft Comput. 9, 236–253 (2005).
Shrivastava, R. K., Singh, S. P. & Kau,r G. Machine learning models for Alzheimer’s disease detection using OASIS data. in Data Analysis for Neurodegenerative Disorders 111–126 (Springer, 2023).
Ghosh, T. et al. A robust distributed deep learning approach to detect Alzheimer’s Disease from MRI images. Mathematics 11, 2633 (2023).
Farshidvard, A., Hooshmand, F. & MirHassani, S. A novel two-phase clustering-based under-sampling method for imbalanced classification problems. Expert Syst. Appl. 213, 119003 (2023).
Mehmood, A., Shahid, F., Khan, R., Ibrahim, M. M. & Zheng, Z. Utilizing siamese 4D-AlzNet and transfer learning to identify stages of Alzheimer’s disease. Neuroscience 545, 69–85 (2024).
Mahmud, T. et al. An explainable AI paradigm for Alzheimer’s diagnosis using deep transfer learning. Diagnostics 14, 345 (2024).
Kırtay, S. & Koçak, M. T. Transfer learning in severity classification in Alzheimer’s: a benchmark comparative study on deep neural networks. Aurum J. Health Sci. 6, 91–108 (2024).
Naqvi, S. M. A., Shabaz, M., Khan, M. A. & Hassan, S. I. Adversarial attacks on visual objects using the fast gradient sign method. J. Grid Comput. 21, 52 (2023).
Yang, X.-S. Firefly algorithm, stochastic test functions and design optimisation. Int. J. Bio-Inspir. Comput. 2, 78–84 (2010).
Tubishat, M. et al. Dynamic salp swarm algorithm for feature selection. Expert Syst. Appl. 164, 113873 (2021).
Yildizdan, G. & Baykan, Ö. K. A novel modified bat algorithm hybridizing by differential evolution algorithm. Expert Syst. Appl. 141, 112949 (2020).
Yang, X.-S. & Deb, S. Cuckoo search via Lévy flights. In Proc. World Congress on Nature & Biologically Inspired Computing (NaBIC) 210–214 (IEEE, 2009).
Mousavirad, S. J., Schaefer, G. & Ebrahimpour-Komleh, H. The human mental search algorithm for solving optimisation problems. in Enabling AI Applications in Data Science 27–47 (Springer International Publishing, 2021).
Karaman, A. et al. Robust real-time polyp detection system design based on YOLO algorithms by optimizing activation functions and hyper-parameters with artificial bee colony (ABC). Expert Syst. Appl. 221, 119741 (2023).
Lu, S. A short survey on computer-aided diagnosis of alzheimer’s disease: unsupervised learning, transfer learning, and other machine learning methods. AI Med. 1, 2–2 (2024).
Ma, H. et al. Classification of Alzheimer’s disease: application of a transfer learning deep Q‐network method. Eur. J. Neurosci. 59, 2118–2127 (2024).
Ali, M. U. et al. Enhancing Alzheimer’s disease diagnosis and staging: a multistage CNN framework using MRI. Front. Psychiatry 15, 1395563 (2024).
Arivalagan, D., Manav, M., & Vignesh, O. Unveiling Alzheimer’s stages through transfer learning and custom CNN layers. In Proc. International Conference on Distributed Computing and Optimization Techniques (ICDCOT) 1–6 (IEEE, 2024).
Acknowledgements
Not applicable.
Author information
Authors and Affiliations
Contributions
Zhisen He: Conceptualization, Methodology, Validation, Investigation, Resources, Writing—original draft, Writing—review and editing, Visualization. Vijay Govindarajan: Software, Data curation, Funding acquisition, Validation, Formal analysis, Investigation. Jing Yang: Conceptualization, Methodology, Software, Writing—review and editing, Visualization. Mahmoud Abdel-Salam: Writing—review and editing, Visualization. Zaffar Ahmed Shaikh: Methodology, Software, Validation, Formal analysis, Writing—review and editing. Lip Yee Por: Investigation, Resources, Writing—original draft, Project administration. Roohallah Alizadehsani: Conceptualization, Software, Formal analysis, Investigation. Ru-San Tan: Methodology, Validation, Visualization, Data curation, Writing—review and editing. U. Rajendra Acharya: Methodology, Validation, Visualization, Data curation, Writing—review and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
He, Z., Govindarajan, V., Yang, J. et al. Automated Alzheimer’s disease detection using active learning model with reinforcement learning and scope loss function. npj Mental Health Res 4, 49 (2025). https://doi.org/10.1038/s44184-025-00158-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s44184-025-00158-2