Abstract
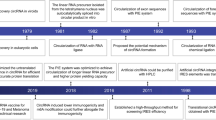
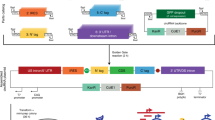
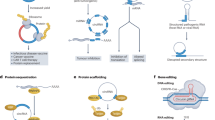
Circular RNAs (circRNAs) are a group of RNA molecules prevalent across various organisms and tissues and characterized by a covalent loop structure. Their unique structure, lacking 5′ and 3′ ends, confers resistance to exonucleases, thereby enhancing their stability compared to linear RNAs. Since the early 2010s, the versatility of circRNAs have been highlighted in applications such as RNA aptamers, guide RNAs and, more recently, SARS-CoV-2 vaccines. Recent advances in rational design, as well as in vitro and in vivo synthesis techniques, underscore the potential for large-scale engineering and production of circRNAs, positioning them as promising candidates for stable and efficient RNA-based therapeutics with minimal immunogenicity. This Review summarizes the guiding principles behind circRNA engineering and development, with a focus on key design elements. We also provide an overview of circRNA advances in disease prevention and treatment. By emphasizing existing limitations and outlining future milestones, this Review offers a translational outlook on circRNAs as an emerging field in biomedicine.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Crooke, S. T., Baker, B. F., Crooke, R. M. & Liang, X. H. Antisense technology: an overview and prospectus. Nat. Rev. Drug Discov. 20, 427–453 (2021).
Xiong, Q. & Zhang, Y. Small RNA modifications: regulatory molecules and potential applications. J. Hematol. Oncol. 16, 64 (2023).
Damase, T. R. et al. The limitless future of RNA therapeutics. Front. Bioeng. Biotech. 9, 628137 (2021).
Zhou, J. & Rossi, J. Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov. 16, 181–202 (2017).
Zhou, J. & Rossi, J. J. Cell-type-specific, aptamer-functionalized agents for targeted disease therapy. Mol. Ther. Nucleic Acids 3, e169 (2014).
Walsh, E. E. et al. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N. Engl. J. Med. 383, 2439–2450 (2020).
Zhu, Y., Zhu, L., Wang, X. & Jin, H. RNA-based therapeutics: an overview and prospectus. Cell Death Dis. 13, 644 (2022).
Ji, P. et al. Expanded expression landscape and prioritization of circular RNAs in mammals. Cell Rep. 26, 3444–3460.e5 (2019).
Wu, W., Zhao, F. & Zhang, J. circAtlas 3.0: a gateway to 3 million curated vertebrate circular RNAs based on a standardized nomenclature scheme. Nucleic Acids Res. 52, D52–D60 (2024).
Memczak, S. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338 (2013).
Hansen, T. B. et al. Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388 (2013).
Ashwal-Fluss, R. et al. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 56, 55–66 (2014).
Li, Z. et al. Exon–intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 22, 256–264 (2015).
Song, R. et al. A novel polypeptide encoded by the circular RNA ZKSCAN1 suppresses HCC via degradation of mTOR. Mol. Cancer 22, 16 (2023).
Fan, H. N. et al. METTL14-mediated m(6)A modification of circORC5 suppresses gastric cancer progression by regulating miR-30c-2-3p/AKT1S1 axis. Mol. Cancer 21, 51 (2022).
Du, J. et al. CircNFIB inhibits tumor growth and metastasis through suppressing MEK1/ERK signaling in intrahepatic cholangiocarcinoma. Mol. Cancer 21, 18 (2022).
Zhao, Q. et al. Targeting mitochondria-located circRNA SCAR alleviates NASH via reducing mROS output. Cell 183, 76–93.e22 (2020).
Huang, D. et al. Tumour circular RNAs elicit anti-tumour immunity by encoding cryptic peptides. Nature 625, 593–602 (2024).
Wang, C. & Liu, H. Factors influencing degradation kinetics of mRNAs and half-lives of microRNAs, circRNAs, lncRNAs in blood in vitro using quantitative PCR. Sci. Rep 12, 7259 (2022).
Nakamoto, K. et al. Chemically synthesized circular RNAs with phosphoramidate linkages enable rolling circle translation. Chem. Commun. 56, 6217–6220 (2020).
Chen, C. Y. & Sarnow, P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 268, 415–417 (1995).
Beaudry, D. & Perreault, J. P. An efficient strategy for the synthesis of circular RNA molecules. Nucleic Acids Res. 23, 3064–3066 (1995).
Wesselhoeft, R. A., Kowalski, P. S. & Anderson, D. G. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun. 9, 2629 (2018).
Lee, K. H. et al. Efficient circular RNA engineering by end-to-end self-targeting and splicing reaction using Tetrahymena group I intron ribozyme. Mol. Ther. Nucleic Acids 33, 587–598 (2023).
Chen, C. et al. A flexible, efficient, and scalable platform to produce circular RNAs as new therapeutics. Preprint at bioRxiv https://doi.org/10.1101/2022.05.31.494115 (2022).
Unti, M. J. & Jaffrey, S. R. Highly efficient cellular expression of circular mRNA enables prolonged protein expression. Cell Chem. Biol. 31, 163–176.e5 (2024).
Qu, S. et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 365, 141–148 (2015).
Jeck, W. R. & Sharpless, N. E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 32, 453–461 (2014).
Wang, X., Jian, W., Luo, Q. & Fang, L. CircSEMA4B inhibits the progression of breast cancer by encoding a novel protein SEMA4B-211aa and regulating AKT phosphorylation. Cell Death Dis. 13, 794 (2022).
Zhang, J. et al. CircRAPGEF5 interacts with RBFOX2 to confer ferroptosis resistance by modulating alternative splicing of TFRC in endometrial cancer. Redox Biol. 57, 102493 (2022).
Jackson, R. J., Hellen, C. U. & Pestova, T. V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127 (2010).
Zhao, J. et al. IRESbase: a comprehensive database of experimentally validated internal ribosome entry sites. Genom. Proteom. Bioinf. 18, 129–139 (2020).
Dresios, J., Chappell, S. A., Zhou, W. & Mauro, V. P. An mRNA–rRNA base-pairing mechanism for translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 13, 30–34 (2006).
Chen, C. K. et al. Structured elements drive extensive circular RNA translation. Mol. Cell 81, 4300–4318.e13 (2021).
Lacerda, R., Menezes, J. & Romao, L. More than just scanning: the importance of cap-independent mRNA translation initiation for cellular stress response and cancer. Cell Mol. Life Sci. 74, 1659–1680 (2017).
Godet, A. C. et al. IRES trans-acting factors, key actors of the stress response. Int. J. Mol. Sci. 20, 924 (2019).
Chen, R. et al. Engineering circular RNA for enhanced protein production. Nat. Biotechnol. 41, 262–272 (2023).
Kameda, S., Ohno, H. & Saito, H. Synthetic circular RNA switches and circuits that control protein expression in mammalian cells. Nucleic Acids Res. 51, e24 (2023).
Ning, H. et al. Rational design of microRNA-responsive switch for programmable translational control in mammalian cells. Nat. Commun. 14, 7193 (2023).
Yang, Y. et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 27, 626–641 (2017).
Csepany, T., Lin, A., Baldick, C. J. Jr. & Beemon, K. Sequence specificity of mRNA N6-adenosine methyltransferase. J. Biol. Chem. 265, 20117–20122 (1990).
Harper, J. E., Miceli, S. M., Roberts, R. J. & Manley, J. L. Sequence specificity of the human mRNA N6-adenosine methylase in vitro. Nucleic Acids Res. 18, 5735–5741 (1990).
Zhou, C. et al. Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep. 20, 2262–2276 (2017).
Zhao, J. et al. Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat. Commun. 10, 2300 (2019).
Chen, Y. G. et al. N6-methyladenosine modification controls circular RNA immunity. Mol. Cell 76, 96–109.e9 (2019).
Chen, J. et al. Pervasive functional translation of noncanonical human open reading frames. Science 367, 1140–1146 (2020).
Ho-Xuan, H. et al. Comprehensive analysis of translation from overexpressed circular RNAs reveals pervasive translation from linear transcripts. Nucleic Acids Res. 48, 10368–10382 (2020).
Jiang, Y., Chen, X. & Zhang, W. Overexpression-based detection of translatable circular RNAs is vulnerable to coexistent linear RNA byproducts. Biochem. Biophys. Res. Commun. 558, 189–195 (2021).
Abe, N. et al. Rolling circle translation of circular RNA in living human cells. Sci. Rep. 5, 16435 (2015).
Wen, S. Y., Qadir, J. & Yang, B. B. Circular RNA translation: novel protein isoforms and clinical significance. Trends Mol. Med. 28, 405–420 (2022).
Dudekula, D. B. et al. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 13, 34–42 (2016).
AbouHaidar, M. G., Venkataraman, S., Golshani, A., Liu, B. & Ahmad, T. Novel coding, translation, and gene expression of a replicating covalently closed circular RNA of 220 nt. Proc. Natl Acad. Sci. USA 111, 14542–14547 (2014).
Fan, X., Yang, Y., Chen, C. & Wang, Z. Pervasive translation of circular RNAs driven by short IRES-like elements. Nat. Commun. 13, 3751 (2022).
Liu, L. et al. Engineering circularized mRNAs for the production of spider silk proteins. Appl. Env. Microb. 88, e0002822 (2022).
Barbosa, C., Peixeiro, I. & Romao, L. Gene expression regulation by upstream open reading frames and human disease. PLoS Genet. 9, e1003529 (2013).
Ryczek, N., Lys, A. & Makalowska, I. The functional meaning of 5′UTR in protein-coding genes. Int. J. Mol. Sci. 24, e24032976 (2023).
Mayr, C. What are 3′ UTRs doing? Cold Spring Harb. Perspect. Biol. 11, a034728 (2019).
Mitschka, S. & Mayr, C. Context-specific regulation and function of mRNA alternative polyadenylation. Nat. Rev. Mol. Cell Biol. 23, 779–796 (2022).
Leppek, K., Das, R. & Barna, M. Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol. 19, 158–174 (2018).
Kozak, M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44, 283–292 (1986).
Qu, L. et al. Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell 185, 1728–1744.e16 (2022).
Holcik, M. & Liebhaber, S. A. Four highly stable eukaryotic mRNAs assemble 3′ untranslated region RNA–protein complexes sharing cis and trans components. Proc. Natl Acad. Sci. USA 94, 2410–2414 (1997).
Carrieri, C. et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491, 454–457 (2012).
Morales-Martinez, M. & Vega, M. I. Role of microRNA-7 (miR-7) in cancer physiopathology. Int. J. Mol. Sci. 23, e23169091 (2022).
Abere, B. et al. Kaposi’s sarcoma-associated herpesvirus-encoded circrnas are expressed in infected tumor tissues and are incorporated into virions. mBio 11, e03027-19 (2020).
Zhang, Y. et al. CircDYM ameliorates depressive-like behavior by targeting miR-9 to regulate microglial activation via HSP90 ubiquitination. Mol. Psychiatry 25, 1175–1190 (2020).
Han, D. et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology 66, 1151–1164 (2017).
Wang, L. et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-β-induced epithelial-mesenchymal transition and metastasis by controlling TIF1γ in non-small cell lung cancer. Mol. Cancer 17, 140 (2018).
Ye, F. et al. circFBXW7 inhibits malignant progression by sponging miR-197-3p and encoding a 185-aa protein in triple-negative breast cancer. Mol. Ther. Nucleic Acids 18, 88–98 (2019).
Rossbach, O. Artificial circular RNA sponges targeting micrornas as a novel tool in molecular biology. Mol. Ther. Nucleic Acids 17, 452–454 (2019).
Liu, X. et al. Synthetic circular RNA functions as a miR-21 sponge to suppress gastric carcinoma cell proliferation. Mol. Ther. Nucleic Acids 13, 312–321 (2018).
Jost, I. et al. Functional sequestration of microRNA-122 from hepatitis C virus by circular RNA sponges. RNA Biol. 15, 1032–1039 (2018).
Muller, S. et al. Synthetic circular miR-21 RNA decoys enhance tumor suppressor expression and impair tumor growth in mice. NAR Cancer 2, zcaa014 (2020).
Lavenniah, A. et al. Engineered circular RNA sponges act as miRNA inhibitors to attenuate pressure overload-induced cardiac hypertrophy. Mol. Ther. 28, 1506–1517 (2020).
Jarlstad Olesen, M. T. & S Kristensen, L. Circular RNAs as microRNA sponges: evidence and controversies. Essays Biochem. 65, 685–696 (2021).
Du, W. W. et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 44, 2846–2858 (2016).
Liu, C. X. & Chen, L. L. Circular RNAs: characterization, cellular roles, and applications. Cell 185, 2016–2034 (2022).
Chen, S. et al. circVAMP3 drives CAPRIN1 phase separation and inhibits hepatocellular carcinoma by suppressing c-Myc translation. Adv. Sci. 9, e2103817 (2022).
Schreiner, S., Didio, A., Hung, L. H. & Bindereif, A. Design and application of circular RNAs with protein-sponge function. Nucleic Acids Res. 48, 12326–12335 (2020).
Abdelmohsen, K. et al. Circular RNAs in monkey muscle: age-dependent changes. Aging 7, 903–910 (2015).
Xu, X. et al. CircRNA inhibits DNA damage repair by interacting with host gene. Mol. Cancer 19, 128 (2020).
Zhao, C. et al. CircFOXO3 protects against osteoarthritis by targeting its parental gene FOXO3 and activating PI3K/AKT-mediated autophagy. Cell Death Dis. 13, 932 (2022).
Pfafenrot, C. et al. Inhibition of SARS-CoV-2 coronavirus proliferation by designer antisense-circRNAs. Nucleic Acids Res. 49, 12502–12516 (2021).
Ren, S. et al. Efficient modulation of exon skipping via antisense circular RNAs. Res. China 6, 0045 (2023).
Wu, N. et al. Silencing mouse circular RNA circSlc8a1 by circular antisense cA-circSlc8a1 induces cardiac hepatopathy. Mol. Ther. 31, 1688–1704 (2023).
Katrekar, D. et al. Efficient in vitro and in vivo RNA editing via recruitment of endogenous ADARs using circular guide RNAs. Nat. Biotechnol. 40, 938–945 (2022).
Yi, Z. et al. Engineered circular ADAR-recruiting RNAs increase the efficiency and fidelity of RNA editing in vitro and in vivo. Nat. Biotechnol. 40, 946–955 (2022).
Liang, R. et al. Prime editing using CRISPR–Cas12a and circular RNAs in human cells. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-02095-x (2024).
Obi, P. & Chen, Y. G. The design and synthesis of circular RNAs. Methods 196, 85–103 (2021).
Beckert, B. & Masquida, B. Synthesis of RNA by in vitro transcription. Meth. Mol. Biol. 703, 29–41 (2011).
Muller, S. & Appel, B. In vitro circularization of RNA. RNA Biol. 14, 1018–1027 (2017).
Deana, A., Celesnik, H. & Belasco, J. G. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature 451, 355–358 (2008).
Petkovic, S. & Muller, S. Synthesis and engineering of circular RNAs. Meth. Mol. Biol. 1724, 167–180 (2018).
Dolinnaya, N. G., Sokolova, N. I., Ashirbekova, D. T. & Shabarova, Z. A. The use of BrCN for assembling modified DNA duplexes and DNA-RNA hybrids; comparison with water-soluble carbodiimide. Nucleic Acids Res. 19, 3067–3072 (1991).
Lohman, G. J., Tabor, S. & Nichols, N. M. DNA ligases. Curr. Protoc. Mol. Biol. 94, 3.14.1–3.14.7 (2011).
Nilsson, M., Antson, D. O., Barbany, G. & Landegren, U. RNA-templated DNA ligation for transcript analysis. Nucleic Acids Res. 29, 578–581 (2001).
Bullard, D. R. & Bowater, R. P. Direct comparison of nick-joining activity of the nucleic acid ligases from bacteriophage T4. Biochem. J. 398, 135–144 (2006).
Chen, H. et al. Preferential production of RNA rings by T4 RNA ligase 2 without any splint through rational design of precursor strand. Nucleic Acids Res. 48, e54 (2020).
Zhao, N. N. et al. Construction of genetically encoded light-up RNA aptamers for label-free and ultrasensitive detection of circRNAs in cancer cells and tissues. Anal. Chem. 95, 8728–8734 (2023).
Puttaraju, M. & Been, M. D. Group I permuted intron–exon (PIE) sequences self-splice to produce circular exons. Nucleic Acids Res. 20, 5357–5364 (1992).
Umekage, S. & Kikuchi, Y. In vivo circular RNA production using a constitutive promoter for high-level expression. J. Biosci. Bioeng. 108, 354–356 (2009).
Li, H. et al. Circular RNA cancer vaccines drive immunity in hard-to-treat malignancies. Theranostics 12, 6422–6436 (2022).
Liu, C. X. et al. RNA circles with minimized immunogenicity as potent PKR inhibitors. Mol. Cell 82, 420–434.e6 (2022).
Guo, S. K. et al. Therapeutic application of circular RNA aptamers in a mouse model of psoriasis. Nat. Biotechnol. https://doi.org/10.1038/s41587-024-02204-4 (2024).
Qiu, Z. et al. Clean-PIE: a novel strategy for efficiently constructing precise circRNA with thoroughly minimized immunogenicity to direct potent and durable protein expression. Preprint at bioRxiv https://doi.org/10.1101/2022.06.20.496777 (2022).
Smathers, C. M. & Robart, A. R. The mechanism of splicing as told by group II introns: ancestors of the spliceosome. Biochim. Biophys. Acta Gene Regul. Mech. 1862, 194390 (2019).
Xu, L., Liu, T., Chung, K. & Pyle, A. M. Structural insights into intron catalysis and dynamics during splicing. Nature 624, 682–688 (2023).
Rupert, P. B. & Ferre-D’Amare, A. R. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature 410, 780–786 (2001).
Chen, X. & Lu, Y. Circular RNA: biosynthesis in vitro. Front. Bioeng. Biotech. 9, 787881 (2021).
Hieronymus, R. & Muller, S. Engineering of hairpin ribozyme variants for RNA recombination and splicing. Ann. NY Acad. Sci. 1447, 135–143 (2019).
Hansen, C. E., Springstubbe, D., Müller, S. & Petkovic, S. in Circular RNAs Methods in Molecular Biology Vol. 12 (eds Dieterich, C. & Baudet, M.-L.) 209–226 (Springer, 2024).
Fantoni, N. Z., El-Sagheer, A. H. & Brown, T. A hitchhiker’s guide to click-chemistry with nucleic acids. Chem. Rev. 121, 7122–7154 (2021).
Kim, Y. S. et al. The RNA ligation method using modified splint DNAs significantly improves the efficiency of circular RNA synthesis. Anim. Cell Syst. 27, 208–218 (2023).
Jaeger, L., Wright, M. C. & Joyce, G. F. A complex ligase ribozyme evolved in vitro from a group I ribozyme domain. Proc. Natl Acad. Sci. USA 96, 14712–14717 (1999).
Wesselhoeft, R. A. et al. RNA circularization diminishes immunogenicity and can extend translation duration in vivo. Mol. Cell 74, 508–520.e4 (2019).
Suzuki, H. et al. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 34, e63 (2006).
Xiao, M. S. & Wilusz, J. E. An improved method for circular RNA purification using RNase R that efficiently removes linear RNAs containing G-quadruplexes or structured 3′ ends. Nucleic Acids Res. 47, 8755–8769 (2019).
Abe, B. T., Wesselhoeft, R. A., Chen, R., Anderson, D. G. & Chang, H. Y. Circular RNA migration in agarose gel electrophoresis. Mol. Cell 82, 1768–1777.e3 (2022).
Niu, D., Wu, Y. & Lian, J. Circular RNA vaccine in disease prevention and treatment. Signal Transduct. Target. Ther. 8, 341 (2023).
Conn, S. J. et al. The RNA binding protein quaking regulates formation of circRNAs. Cell 160, 1125–1134 (2015).
Li, X. et al. A unified mechanism for intron and exon definition and back-splicing. Nature 573, 375–380 (2019).
Costello, A., Lao, N. T., Barron, N. & Clynes, M. Continuous translation of circularized mRNA improves recombinant protein titer. Metab. Eng. 52, 284–292 (2019).
Schmidt, C. A., Giusto, J. D., Bao, A., Hopper, A. K. & Matera, A. G. Molecular determinants of metazoan tricRNA biogenesis. Nucleic Acids Res. 47, 6452–6465 (2019).
Litke, J. L. & Jaffrey, S. R. Highly efficient expression of circular RNA aptamers in cells using autocatalytic transcripts. Nat. Biotechnol. 37, 667–675 (2019).
Nielsen, A. F. et al. Best practice standards for circular RNA research. Nat. Meth. 19, 1208–1220 (2022).
Daros, J. A. Production of circular recombinant RNA in Escherichia coli using viroid scaffolds. Meth. Mol. Biol. 2323, 99–107 (2021).
Yuan, Q. et al. CircRNA DICAR as a novel endogenous regulator for diabetic cardiomyopathy and diabetic pyroptosis of cardiomyocytes. Signal Transduct. Target. Ther. 8, 99 (2023).
Zhu, Y. et al. Circ-Ddx60 contributes to the antihypertrophic memory of exercise hypertrophic preconditioning. J. Adv. Res. 46, 113–121 (2023).
Zhou, Z. et al. CircDYM attenuates microglial apoptosis via CEBPB/ZC3H4 axis in LPS-induced mouse model of depression. Int. J. Biol. Macromol. 254, 127922 (2024).
Bulcha, J. T., Wang, Y., Ma, H., Tai, P. W. L. & Gao, G. Viral vector platforms within the gene therapy landscape. Signal Transduct. Target. Ther. 6, 53 (2021).
Gao, Y., Wang, J. & Zhao, F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 16, 4 (2015).
Gao, Y., Zhang, J. & Zhao, F. Circular RNA identification based on multiple seed matching. Brief. Bioinform. 19, 803–810 (2017).
Amaya, L. et al. Pathways for macrophage uptake of cell-free circular RNAs. Mol. Cell 84, 2104–2118.e6 (2024).
Ngo, L. H. et al. Nuclear export of circular RNA. Nature 627, 212–220 (2024).
Cao, S. M. et al. Altered nucleocytoplasmic export of adenosine-rich circRNAs by PABPC1 contributes to neuronal function. Mol. Cell 84, 2304–2319.e8 (2024).
Chaudhary, N., Weissman, D. & Whitehead, K. A. mRNA vaccines for infectious diseases: principles, delivery and clinical translation. Nat. Rev. Drug Discov. 20, 817–838 (2021).
Pardi, N., Hogan, M. J., Porter, F. W. & Weissman, D. mRNA vaccines — a new era in vaccinology. Nat. Rev. Drug Discov. 17, 261–279 (2018).
Rosa, S. S., Prazeres, D. M. F., Azevedo, A. M. & Marques, M. P. C. mRNA vaccines manufacturing: challenges and bottlenecks. Vaccine 39, 2190–2200 (2021).
Enuka, Y. et al. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 44, 1370–1383 (2016).
Chen, Y. G. et al. Sensing self and foreign circular RNAs by intron identity. Mol. Cell 67, 228–238.e5 (2017).
Seephetdee, C. et al. A circular mRNA vaccine prototype producing VFLIP-X spike confers a broad neutralization of SARS-CoV-2 variants by mouse sera. Antivir. Res. 204, 105370 (2022).
Wan, J. et al. Circular RNA vaccines with long-term lymph node-targeting delivery stability after lyophilization induce potent and persistent immune responses. mBio 15, e0177523 (2024).
Zhu, F. et al. Development of a novel circular mRNA vaccine of six protein combinations against Staphylococcus aureus. J. Biomol. Struct. Dyn. 41, 10525–10545 (2023).
Amaya, L. et al. Circular RNA vaccine induces potent T cell responses. Proc. Natl Acad. Sci. USA 120, e2302191120 (2023).
Vavilis, T. et al. mRNA in the context of protein replacement therapy. Pharmaceutics 15, 166 (2023).
Perez-Garcia, C. G. et al. Development of an mRNA replacement therapy for phenylketonuria. Mol. Ther. Nucleic Acids 28, 87–98 (2022).
Anttila, V. et al. Direct intramyocardial injection of VEGF mRNA in patients undergoing coronary artery bypass grafting. Mol. Ther. 31, 866–874 (2023).
Zhang, M. et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat. Commun. 9, 4475 (2018).
Song, J. et al. A novel protein encoded by ZCRB1-induced circHEATR5B suppresses aerobic glycolysis of GBM through phosphorylation of JMJD5. J. Exp. Clin. Cancer Res. 41, 171 (2022).
Schaff, L. R. & Mellinghoff, I. K. Glioblastoma and other primary brain malignancies in adults: a review. JAMA 329, 574–587, (2023).
Liu, B. et al. Cytoskeleton remodeling mediated by circRNA-YBX1 phase separation suppresses the metastasis of liver cancer. Proc. Natl Acad. Sci. USA 120, e2220296120 (2023).
Jiang, T. et al. A novel protein encoded by circMAPK1 inhibits progression of gastric cancer by suppressing activation of MAPK signaling. Mol. Cancer 20, 66 (2021).
Wang, X. et al. CircURI1 interacts with hnRNPM to inhibit metastasis by modulating alternative splicing in gastric cancer. Proc. Natl Acad Sci USA 118, e2012881118 (2021).
Zhang, Y. et al. CircDIDO1 inhibits gastric cancer progression by encoding a novel DIDO1-529aa protein and regulating PRDX2 protein stability. Mol. Cancer 20, 101 (2021).
Guo, Z., Zhang, Y., Xu, W., Zhang, X. & Jiang, J. Engineered exosome-mediated delivery of circDIDO1 inhibits gastric cancer progression via regulation of MiR-1307-3p/SOCS2 axis. J. Transl. Med. 20, 326 (2022).
Hu, F. et al. Vimentin binds to a novel tumor suppressor protein, GSPT1-238aa, encoded by circGSPT1 with a selective encoding priority to halt autophagy in gastric carcinoma. Cancer Lett. 545, 215826 (2022).
Wei, S. et al. The circRNA circPTPRA suppresses epithelial-mesenchymal transitioning and metastasis of NSCLC cells by sponging miR-96-5p. eBiomedicine 44, 182–193 (2019).
Wang, T. et al. A novel protein encoded by circASK1 ameliorates gefitinib resistance in lung adenocarcinoma by competitively activating ASK1-dependent apoptosis. Cancer Lett. 520, 321–331 (2021).
Feng, Z. et al. An in vitro-transcribed circular RNA targets the mitochondrial inner membrane cardiolipin to ablate EIF4G2+/PTBP1+ pan-adenocarcinoma. Nat. Cancer 5, 30–46 (2024).
Chen, Q. et al. CircRNA cRAPGEF5 inhibits the growth and metastasis of renal cell carcinoma via the miR-27a-3p/TXNIP pathway. Cancer Lett. 469, 68–77 (2020).
Li, F. et al. A peptide CORO1C-47aa encoded by the circular noncoding RNA circ-0000437 functions as a negative regulator in endometrium tumor angiogenesis. J. Biol. Chem. 297, 101182 (2021).
Shen, X. et al. Hsa_circ_0000437 promotes pathogenesis of gastric cancer and lymph node metastasis. Oncogene 41, 4724–4735 (2022).
Wang, C. et al. Characterization of distinct circular RNA signatures in solid tumors. Mol. Cancer 21, 63 (2022).
Yan, L., Zheng, M. & Wang, H. CircularR. N. A. hsa_circ_0072309 inhibits proliferation and invasion of breast cancer cells via targeting miR-492. Cancer Manag. Res. 11, 1033–1041 (2019).
Guo, X. et al. Circular RNA hsa_circ_0072309 inhibits the proliferation, invasion and migration of gastric cancer cells via inhibition of PI3K/AKT signaling by activating PPARγ/PTEN signaling. Mol. Med. Rep. 23, 349 (2021).
Ji, J. et al. Downregulation of circLIFR exerts cancer-promoting effects on hepatocellular carcinoma in vitro. Front. Genet. 13, 986322 (2022).
Zhang, H. et al. CircLIFR synergizes with MSH2 to attenuate chemoresistance via MutSalpha/ATM-p73 axis in bladder cancer. Mol. Cancer 20, 70 (2021).
Zhang, X. Q., Song, Q. & Zeng, L. X. Circulating hsa_circ_0072309, acting via the miR-100/ACKR3 pathway, maybe a potential biomarker for the diagnosis, prognosis, and treatment of brain metastasis from non-small-cell lung cancer. Cancer Med. 12, 18005–18019 (2023).
Zhang, J. et al. Comprehensive profiling of circular RNAs with nanopore sequencing and CIRI-long. Nat. Biotechnol. 39, 836–845 (2021).
Xin, R. et al. isoCirc catalogs full-length circular RNA isoforms in human transcriptomes. Nat. Commun. 12, 266 (2021).
Chu, Y. et al. A 5′ UTR language model for decoding untranslated regions of mRNA and function predictions. Nat. Mach. Intell. 6, 449–460 (2024).
Sumi, S., Hamada, M. & Saito, H. Deep generative design of RNA family sequences. Nat. Meth. 21, 435–443 (2024).
Loan Young, T., Chang Wang, K., James Varley, A. & Li, B. Clinical delivery of circular RNA: lessons learned from RNA drug development. Adv. Drug Deliv. Rev. 197, 114826 (2023).
Wu, W., Zhang, J., Cao, X., Cai, Z. & Zhao, F. Exploring the cellular landscape of circular RNAs using full-length single-cell RNA sequencing. Nat. Commun. 13, 3242 (2022).
Zhou, Z. et al. CIRI-deep enables single-cell and spatial transcriptomic analysis of circular RNAs with deep learning. Adv. Sci. 11, e2308115 (2024).
Liu, C. X. et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell 177, 865–880.e21 (2019).
Ren, L. et al. Mechanisms of circular RNA degradation. Commun. Biol. 5, 1355 (2022).
Li, J. et al. A nanodrug system overexpressed circRNA_0001805 alleviates nonalcoholic fatty liver disease via miR-106a-5p/miR-320a and ABCA1/CPT1 axis. J. Nanobiotechnol. 19, 363 (2021).
Xu, X. et al. Multifunctional envelope-type siRNA delivery nanoparticle platform for prostate cancer therapy. ACS Nano 11, 2618–2627 (2017).
Zielonka, J. et al. Mitochondria-targeted triphenylphosphonium-based compounds: syntheses, mechanisms of action, and therapeutic and diagnostic applications. Chem. Rev. 117, 10043–10120 (2017).
Gulati, S. et al. Metal–organic frameworks (MOFs) as effectual diagnostic and therapeutic tools for cancer. J. Mater. Chem. B 11, 6782–6801 (2023).
Zhang, Y. et al. Exosomal circRNA as a novel potential therapeutic target for multiple myeloma-related peripheral neuropathy. Cell Signal. 78, 109872 (2021).
Mao, G. et al. Exosome-transported circRNA_0001236 enhances chondrogenesis and suppress cartilage degradation via the miR-3677-3p/Sox9 axis. Stem Cell Res. Ther. 12, 389 (2021).
Han, Y., Liu, Y., Zhang, B. & Yin, G. Exosomal circRNA 0001445 promotes glioma progression through miRNA-127-5p/SNX5 pathway. Aging 13, 13287–13299 (2021).
Fan, L. et al. Exosome-based mitochondrial delivery of circRNA mSCAR alleviates sepsis by orchestrating macrophage activation. Adv. Sci. 10, e2205692 (2023).
Zhang, J. et al. Therapeutic potential of exosomal circRNA derived from synovial mesenchymal cells via targeting circEDIL3/miR-485-3p/PIAS3/STAT3/VEGF functional module in rheumatoid arthritis. Int. J. Nanomed. 16, 7977–7994 (2021).
Hansen, T. B. et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 30, 4414–4422 (2011).
Pan, Z. et al. MicroRNA-1224 splicing circularRNA-Filip1l in an Ago2-dependent manner regulates chronic inflammatory pain via targeting Ubr5. J. Neurosci. 39, 2125–2143 (2019).
Park, O. H. et al. Endoribonucleolytic cleavage of m6A-containing RNAs by RNase P/MRP complex. Mol. Cell 74, 494–507.e8 (2019).
Zhang, L. et al. The role of N6-methyladenosine (m6A) modification in the regulation of circRNAs. Mol. Cancer 19, 105 (2020).
Guo, Y. et al. Circ3823 contributes to growth, metastasis and angiogenesis of colorectal cancer: involvement of miR-30c-5p/TCF7 axis. Mol. Cancer 20, 93 (2021).
Fischer, J. W., Busa, V. F., Shao, Y. & Leung, A. K. L. Structure-mediated RNA decay by UPF1 and G3BP1. Mol. Cell 78, 70–84.e6 (2020).
Guo, Y. et al. A diet high in sugar and fat influences neurotransmitter metabolism and then affects brain function by altering the gut microbiota. Transl. Psychiatry 11, 328 (2021).
Yang, L. et al. Extracellular vesicle-mediated delivery of circular RNA SCMH1 promotes functional recovery in rodent and nonhuman primate ischemic stroke models. Circulation 142, 556–574 (2020).
Yang, Y. et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J. Natl Cancer Inst. 110, 304–315 (2018).
Zhang, M. et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene 37, 1805–1814 (2018).
Xia, X. et al. A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent kinase-1. Mol. Cancer 18, 131 (2019).
Liu, Z. et al. CircRNA-5692 inhibits the progression of hepatocellular carcinoma by sponging miR-328-5p to enhance DAB2IP expression. Cell Death Dis. 10, 900 (2019).
Fang, J. et al. A novel circular RNA, circFAT1(e2), inhibits gastric cancer progression by targeting miR-548g in the cytoplasm and interacting with YBX1 in the nucleus. Cancer Lett. 442, 222–232 (2019).
Zeng, W., Liu, Y., Li, W. T., Li, Y. & Zhu, J. F. CircFNDC3B sequestrates miR-937-5p to derepress TIMP3 and inhibit colorectal cancer progression. Mol. Oncol. 14, 2960–2984 (2020).
Pan, Z. et al. A novel protein encoded by circFNDC3B inhibits tumor progression and EMT through regulating Snail in colon cancer. Mol. Cancer 19, 71 (2020).
Liang, Z. X. et al. A novel NF-kappaB regulator encoded by circPLCE1 inhibits colorectal carcinoma progression by promoting RPS3 ubiquitin-dependent degradation. Mol. Cancer 20, 103 (2021).
Wang, L. et al. A novel tumour suppressor protein encoded by circMAPK14 inhibits progression and metastasis of colorectal cancer by competitively binding to MKK6. Clin. Transl. Med. 11, e613 (2021).
Garikipati, V. N. S. et al. Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat. Commun. 10, 4317 (2019).
Peng, F. et al. circRNA_010383 acts as a sponge for miR-135a, and its downregulated expression contributes to renal fibrosis in diabetic nephropathy. Diabetes 70, 603–615 (2021).
Liu, H. et al. Invasion-related circular RNA circFNDC3B inhibits bladder cancer progression through the miR-1178-3p/G3BP2/SRC/FAK axis. Mol. Cancer 17, 161 (2018).
Lu, Q. et al. Circular RNA circSLC8A1 acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer progression via regulating PTEN. Mol. Cancer 18, 111 (2019).
Merrick, W. C. & Pavitt, G. D. Protein synthesis initiation in eukaryotic cells. Cold Spring Harb. Perspect. Biol. 10, a033092 (2018).
Rozen, F. et al. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol. Cell. Biol. 10, 1134–1144 (1990).
Johannes, G. & Sarnow, P. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA 4, 1500–1513 (1998).
Prevot, D., Darlix, J. L. & Ohlmann, T. Conducting the initiation of protein synthesis: the role of eIF4G. Biol. Cell 95, 141–156 (2003).
Komar, A. A. & Hatzoglou, M. Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle 10, 229–240 (2011).
Lee, K. M., Chen, C. J. & Shih, S. R. Regulation mechanisms of viral IRES-driven translation. Trends Microbiol. 25, 546–561 (2017).
Abdullah, S. W., Wu, J., Wang, X., Guo, H. & Sun, S. Advances and breakthroughs in IRES-directed translation and replication of picornaviruses. mBio 14, e0035823 (2023).
Acknowledgements
This work was supported by grants from the National Key R&D Project (2021YFA1300500 and 2021YFA1302000) and the National Natural Science Foundation of China (32025009, 32130020 and 32200530).
Author information
Authors and Affiliations
Contributions
F.Z. conceived the project. X.C. and Z.C conducted the initial literature search. X.C., J.Z. and F.Z. wrote the manuscript synopsis. All authors contributed to writing and reviewing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Bioengineering thanks Samie Jaffrey and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cao, X., Cai, Z., Zhang, J. et al. Engineering circular RNA medicines. Nat Rev Bioeng 3, 270–287 (2025). https://doi.org/10.1038/s44222-024-00259-1
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s44222-024-00259-1
This article is cited by
-
CircCCT2/miR-146a-5p/IRAK1 axis promotes the development of head and neck squamous cell carcinoma
BMC Cancer (2025)
-
RNA chemistry and therapeutics
Nature Reviews Drug Discovery (2025)
-
Circular RNA discovery with emerging sequencing and deep learning technologies
Nature Genetics (2025)