Abstract
Despite its importance in health, the female intimate skin microbiome remains understudied. In this Isala study we explored microbial dispersal across intimate body sites, including the vagina, groins, breast and mouth. Microbial similarity correlated with physical proximity, suggesting dispersal influenced by hygiene or sexual activity. Notably, lactobacilli were unexpectedly abundant on breast skin. These findings highlight the need for research into microbiome dynamics and their implications for women’s health.
Similar content being viewed by others
Introduction
The human skin is the largest epithelial surface for microbial interaction and a key barrier influenced by endogenous and exogenous factors1,2,3. Its complex topography and multiple layers contribute to a highly variable and phylogenetically diverse microbiome4,5. Most skin typical bacteria reside on the epidermis, but they are also present in deeper layers2,6. Different skin regions host distinct microbial communities: moist areas such as the groin favor Staphylococcus spp. and Corynebacterium spp., sebaceous regions such as forehead are rich in Cutibacterium spp., and dry areas such as forearm and elbow exhibit greater diversity, with taxa such as Corynebacterium spp. and Proteobacterium spp becoming more dominant3,4,7,8.
Disruptions in the skin microbiota composition are linked to inflammatory skin conditions such as acne9,10, psoriasis11, and atopic dermatitis9,12,13. The microbiome plays a crucial role in skin barrier integrity14, modulating inflammatory responses7, wound healing, epidermal differentiation, and tight junction enhancement2,3,15. While lactobacilli are widely recognized as beneficial for gut and vaginal health, their role in skin health is only recently explored14,16,17,18,19. Though the skin’s nutrient-poor and oxygen-rich environment is unfavorable for lactobacilli, reduced levels have been associated with skin diseases in different observational studies16,20,21,22. As the skin is constantly exposed to external microbes, lactobacilli detected on the skin in observational studies may be transient members. They could be transferred from other body sites where they are abundant, such as the vagina23,24. Lactobacilli are also early colonizers of the skin upon vaginal birth16,17, but it is not yet well understood how long they persist. Studies linking vaginal and skin microbiomes of mother to children and within the same (female) individual are needed to confirm such dispersal patterns, but are currently lacking.
Although the vaginal microbiome has gained research interest in relation to pregnancy, fertility, and infections, women remain underrepresented in scientific studies25,26. Gender biases in drug testing and clinical trials persist, leading to gaps in knowledge about the female skin microbiome. Although men and women have distinct skin characteristics—women generally have thinner skin, lower pH, and less sweat production27,28,29—most skin microbiome studies do not distinguish between genders. Additionally, differences in hygiene practices contribute to a more diverse female skin microbiome28,30. Despite this, research rarely specifies whether samples come from men or women, instead summarizing data by body site.
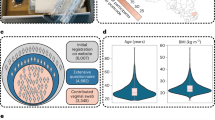
The Isala citizen-science program addressed in its first phase (n = 3345) an important female microbiome knowledge gap by mainly investigating the vaginal microbiome via self-sampling24. Here, we report on the second phase of the Isala project, where women also donated samples of specific intimate skin sites. A subset of 293 participants, selected based on hormone intake, sexual activity, and contraceptive use, participated. During the luteal phase of their menstrual cycle, they provided saliva (data already reported)31; and additional samples reported here: a vaginal swab and skin swabs from the mouth area, breasts, and groin, as well as answered a large survey (Fig. 1a). An overview of cohort selection and (intimate) hygiene characteristics can be found in Supplementary Figs. 1 and 2. General cohort characteristics are described31.
a Study set-up, b Barplot of the microbiome of participant 1, ordered from the vaginal microbiome to the groin skin, breast skin, skin of the mouth and finally the salivary microbiome, c T-sne plot with the trajectory of participant 1 (age: 21, BMI: 17.5) and 2 (age: 38, BMI: 23.3) in the intimate microbiome space, d Barplot of the microbiome of participant 2, ordered as for participant 1, e T-sne plot of the intimate female microbiome space colored for the dominant subgenus at ASV level and clustered based on body site, f T-sne plot of the intimate female microbiome space colored for the second dominant subgenus at ASV level, g t-sne plot of the intimate female microbiome space colored for the relative abundance of the dominant subgenus at ASV level, h Heatmap showing the distance between the respective microbiome sites at ASV level, i Upset plot representing the number of ASV’s found in one or multiple of the sampled body sites, the set size shows the total number of ASV’s found for each body site.
The bacterial DNA of the vaginal, groin, mouth (not the lips) and breast (not the nipple) skin swabs and saliva samples, collected during the luteal phase, was then analysed using the V4 region of the 16S rRNA gene to assess the microbiome of each individual’s intimacy related body site. The V4 region was still selected here as it belongs to the target variable region most optimal for vaginal samples32. We aimed to investigate the dispersal of vaginal-associated Lactobacillus taxa across body sites and were less focused on achieving a comprehensive characterization of the skin microbiome sites. In total 1198 samples passed quality control, resulting in 22 million high-quality V4 amplicon paired reads (Supplementary Fig. 3). Similarly to the vaginal microbiome reads of the first phase of Isala24, the reads were classified up to subgenus level with additional subgenera classification of the genus Lactobacillus24. An overview of the microbiome of each site for all participants can be found in Supplementary Fig. 4. The vaginal microbiome was mainly dominated by Lactobacillus crispatus (54%) and Lactobacillus iners (26%), showing that in the vaginal microbiome, most participants (72%, 178/247 participants) are dominated by the same ASV in their vaginal microbiome of this study phase as during the first phase of the Isala study24. All skin sites were mainly dominated by Staphylococcus sp. and Corynebacterium sp., which is consistent with the skin microbiome of moist skin sites. Interestingly, Cutibacterium spp. were detected in only eight skin samples (3%), with a low average relative abundance (0.15%) given the sebaceous nature of the skin around the mouth and breasts3,4. However, this low abundance can be explained by the study design as it is known that the V4 region fails to properly capture Cutibacterium spp.9,33. The salivary microbiome of this cohort was a conserved community of 12 genera, mainly consisting of Streptococcus, Veillonella and Prevotella, as previously reported by Cauwenberghs et al.31.
We then mapped the community structure of all body sites in a two-dimensional space using t-distributed stochastic neighbour embedding (t-SNE) at ASV level (Fig. 1e)34. Samples clustered on site, which is in accordance with previous findings2,35. When plotting the relative abundance of the top (sub)genus on ASV level, we mainly observed a high relative abundance of a single top (sub)genus in the vaginal microbiome, while the salivary microbiome had an overall lower average relative abundance (Fig. 1g), revealing a core salivary microbiome31. To uncover insights into the dispersal of microbiota through different body sites of one participant, the different samples of two random participants, one with a vaginal microbiome dominated by L. crispatus and one dominated by L. iners, were visualized throughout the microbiome space as examples (Fig. 1b–d). The samples of a single participant were placed on a trajectory through the microbiome space showing the dispersal of more site-specific taxa to nearby body sites. For example, the vaginal microbiome of participant 1 was almost singularly dominated by L. crispatus, which resulted in the immense dispersal of L. crispatus to the groin skin, with a relative abundance of more than 70%, confirmed at ASV level. This almost resembled a more typical vaginal microbiome, with only little Staphylococcus and Corynebacterium present. The groin skin of participant 2 showed less resemblance to her vaginal microbiome, which was dominated by L. iners, but still contained more than 20% of both L. iners and L. jensenii of which the same ASV’s are found in her vaginal microbiome. The microbiome of the skin of the mouth of participant 2 was more similar to the skin microbiome of the breasts, while the mouth skin of participant 1 contained more typical salivary bacteria. These data suggest more dispersal of vaginal fluid to the groin, and the saliva to the skin of the mouth, in participant 1 compared to participant 2, although it remains to be explored whether these ASV’s found on the skin sites are from the same strains as in the vagina or saliva. As such, strain-level resolution is needed to be certain of dispersal, but is beyond the scope of the present study.
To further substantiate possible dispersal to nearby sites, the distance between the ASV’s of the microbiome of each site was determined (Fig. 1h). The distance between microbiomes correlated with physical distance, following a ‘vertical flow’ across body sites; from the vagina to the groin skin, then to the breast skin, the skin around the mouth, and finally, the salivary microbiome. Microbiome (bray-curtis) distances to the vaginal sample were, in order of smallest to largest, the groin skin, breast skin, mouth skin, and finally saliva. A Wilcoxon rank sum test demonstrated that the differences between these distances were significant for each comparison of pairwise distances. This indicate a possible locality-related effect of microbiome similarity, and that the skin microbiome around the mouth is strongly affected by the saliva. This was visible in the number of ASV’s on subgenus level overlapping between sites (Fig. 1i, Supplementary Table 1). The groin skin was an overlap between the vaginal microbiome and skin typical microbiome taxa, while the skin around the mouth showed an overlap between the salivary microbiome and the typical skin microbiome taxa. Unexpectedly, the skin around the breasts contained a notable amount of typical vaginal lactobacilli, with an average relative abundance of 3% on the breasts, compared to an average abundance of 1.9% on the skin around the mouth (Supplementary Fig. 4). We hypothesize a possible distribution of vaginal lactobacilli to other body sites influenced by sexual activity and (intimate) hygiene practices, although the lactobacilli on the breasts could also be endogenously enriched for example due to increased hormone levels and oestrogen receptors in breasts36, which are factors known to stimulate lactobacilli in the vagina. While no significant associations between hygiene habits and recent sexual activity and the skin microbiome sites in our cohort were found (Supplementary Figs 5 and 6), the elevated abundance of lactobacilli on intimate sites is in accordance with Howard B. et al. who found more Lactobacillus sp. on the buttocks of women than on the face and forearm37. Lactobacilli were also already found on the female hands, hinting towards dispersal of the vaginal microbiome38. A possible dispersal due to sexual activity has also been observed in cohabiting couples, where Lactobacillaceae were found on the inner thighs of male partners39. This provides possibly new biological insights in bacterial dispersal, but could also be relevant in sexual assault cases, where the microbiome has recently gained interest as biological evidence40,41,42.
These findings hint towards a distinct female intimate microbiome space beyond the vagina, where vaginal-associated bacteria are present on intimate skin sites. We hypothesize that his dynamic ecosystem could be shaped by hormones, hygiene, and sexual behaviors, forming a continuous microbiome network that may potentially contribute to skin health and microbial balance, although the current set-up of this study is limited in its observational nature and thus fails to uncover these underlying shaping factors. Further research with a more targeted and longitudinal approach is needed to elucidate microbial dispersal, particularly the role of lactobacilli in relation to sexual habits and estrogen levels. Recognizing this space as a unique entity highlights the possibility for targeted studies on its composition, stability, and function. As the dispersal of lactobacilli across body sites has potential implications for microbiome-based therapeutics, infection prevention, and forensic science. Integrating this concept into future research will enhance our understanding of the role of lactobacilli in health and disease beyond the vaginal microbiome.
Methods
Study cohort and data collection
The study was approved by the Ethical Committee of the Antwerp University Hospital/University of Antwerp (B300201942076) and registered online at ClinicalTrials.gov on 2020-03-01 with the unique identifier NCT04319536. This study was conducted in accordance with the ethical principles of the Declaration of Helsinki. Two hundred ninety-three participants were selected from the larger cohort of 3345 women based on hormone intake (no hormones, IUD, combination pill), condom usage (yes, no), frequency of sexual contact (very frequent, frequent, seldom, never) and number of partners (multiple, one, none). Written informed consent was obtained for all participants, in which was stated that participants could be contacted again for follow-up studies. The selection of participants were contacted again and agreed to participate to continue in a second phase where over the course of two menstrual cycles samples were collected. Participants collected vaginal swabs at three timepoints in one menstrual cycle. During the luteal phase of the first menstrual cycle, they also swabbed the skin around the mouth, not including the lips, around the breasts, not including the nipples, and of the groin and collected a saliva sample. Participants were asked to take the samples first thing in the morning, before brushing teeth and were asked to not have showered or put on body cream in the 12 h before. First vaginal swabs were taken, then the skin swabs in the order of mouth to groin and finally the saliva sample. They were asked to wash hands between sampling. Two hundred fifty-eight participants collected all skin samples, 247 participants provided saliva samples, and 255 participants provided vaginal swabs (Supplementary Fig 3). A hundred seventy five participants provided samples of all sites that passed quality control and were used in further analyses. All samples are stored via the Biobank Antwerp, Antwerp, Belgium; ID: BE 71030031000.
Sample collection saliva and skin swabs
Participants provided saliva samples by spitting into a sterile plastic cup and twisting the eNAT® swab in the saliva. For the sampling of the breast skin (the area surrounding the nipples) (n = 258) eNAT® swabs (Copan, Brescia, Italy) were used for microbiome profiling. Participants moistened the swabs with a sterile pre-moisture buffer (50 mM Tris buffer [pH 7,6], 1 mM EDTA [pH 8,0], and 0,5% Tween-20) and gently rubbed the area for 30 s to collect sufficient biomass. The skin of the area around the mouth (n = 260) and groin (n = 258) the participants moisturized the eNAT® swab with 10x PBS and rubbed it for 30 s on a 20 cm² area.
Sample collection and processing of vaginal swabs
The vaginal swabs were collected and processed as described in Lebeer et al.24. 16S rRNA amplicon sequencing and quality control was performed as described in Lebeer et al.24. Analysis was performed using tidytacos43. All samples were sequenced using the V4 region of the 16S rRNA gene, selected for its optimal performance in profiling vaginal microbiomes. While we acknowledge that the V4 region does not fully capture certain skin-associated taxa, our primary aim was to investigate the dispersal of vaginal-associated lactobacilli across body sites.
Statistical analyses
Samples were aggregated from the ASV level to the genus level. Samples from the five sites were embedded together into a common space using t-SNE34,44, using bray-curtis as a distance metric. Distances were calculated between pairs of sites within each participant. Statistical analyses used to determine the associations between microbial community composition and survey data were performed as described in Lebeer et al.24.
Data availability
The sequencing data is available at the European Nuceotide Archive under bioproject PRJEB86101 and PRJEB55042.
Code availability
This paper does not report original code. The in-house amplicon data processing toolkit used is available at https://github.com/SWittouck/tidyamplicons. The tool to perform differential abundance with multiple different testing tools is available at https://github.com/thiesgehrmann/multidiffabundance. All other scripts used for data processing and analysis are hosted on GitHub and accessible at https://github.com/LebeerLab/Citizen-science-map-of-the-vaginal-microbiome.
References
Gallo, R. L. Human skin is the largest epithelial surface for interaction with microbes. J. Investig. Dermatol. 137, 1213–1214 (2017).
Grice, E. A. & Segre, J. A. The skin microbiome. Nat. Rev. Microbiol. y 9, 244–253 (2011).
Skowron, K. et al. Human skin microbiome: impact of intrinsic and extrinsic factors on skin microbiota. Microorganisms 9, 543 (2021).
Grice, E. A. et al. Topographical and temporal diversity of the human skin microbiome. Science 324, 1190–1192 (2009).
Cundell, A. M. Microbial ecology of the human skin. Microbiol. Ecol. 76, 113–120 (2018).
Yousef, H. Alhajj, M. Fakoya, A. O. & Sharma, S. Anatomy, Skin (Integument), Epidermis (StatPearls Publishing 2024).
Belkaid, Y. & Segre, J. A. Dialogue between skin microbiota and immunity. Science ((1979)) 346, 954–959 (2014).
Delanghe, L. et al. The inner elbow skin microbiome contains Lactobacillus among its core taxa and varies with age, season and lifestyle. Microbiome Res. Rep. 3, 43 (2024).
Chen, Y., Knight, R. & Gallo, R. L. Evolving approaches to profiling the microbiome in skin disease. Front. Immunol. 14, 1151527 (2023).
Hou, K. et al. Microbiota in health and diseases. Sig. Transduct. Target. Ther. 7, 135 (2022).
Yerushalmi, M., Elalouf, O., Anderson, M. & Chandran, V. The skin microbiome in psoriatic disease: a systematic review and critical appraisal. J Transl. Autoimmun. 2, 100009 (2019).
Zeeuwen, P. L. J. M., Kleerebezem, M., Timmerman, H. M. & Schalkwijk, J. Microbiome and skin diseases. Curr. Opin. Allergy. Clin. Immunol. 13, 514–520 (2013).
Delanghe, L. et al. Mild atopic dermatitis is characterized by increase in non-staphylococcus pathobionts and loss of specific species. Sci. Rep. 14, 23659 (2024).
Jung, Y. O. et al. Lysates of a probiotic, lactobacillus rhamnosus, can improve skin barrier function in a reconstructed human epidermis model. Int. J. Mol. Sci. 20, 4289 (2019).
Bäsler, K. et al. The role of tight junctions in skin barrier function and dermal absorption. J. of Control. Release 242, 105–118 (2016).
Lebeer, S. et al. Selective targeting of skin pathobionts and inflammation with topically applied lactobacilli. Cell. Rep. Med. 3, 100521 (2022).
Delanghe, L. et al. The role of lactobacilli in inhibiting skin pathogens. Biochem. Soc. Trans. 49, 617–627 (2021).
Butler, É, Lundqvist, C. & Axelsson, J. Lactobacillus reuteri DSM 17938 as a novel topical cosmetic ingredient: a proof of concept clinical study in adults with atopic dermatitis. Microorganisms 8, 1026 (2020).
Blanchet-Réthoré, S. et al. Effect of a lotion containing the heat-treated probiotic strain Lactobacillus johnsonii NCC 533 on Staphylococcus aureus colonization in atopic dermatitis. Clin. Cosmet. Investig. Dermatol. 10, 249–257 (2017).
Olejniczak-Staruch, I. et al. Alterations of the skin and gut microbiome in psoriasis and psoriatic arthritis. Int. J. Mol. Sci. 22, 3998 (2021).
Wang, L. et al. Amplicon-based sequencing and co-occurence network analysis reveals notable differences of microbial community structure in healthy and dandruff scalps. BMC Genomics 23, 312 (2022).
Sánchez-Pellicer, P. et al. Rosacea, microbiome and probiotics: the gut-skin axis. Front. Microbiol. 14, 1323644 (2023).
Glatthardt, T., Lima, R. D., de Mattos, R. M. & Ferreira, R. B. R. Microbe Interactions within the Skin Microbiome. Antibiotics 13, 49 (2024).
Lebeer, S. et al. A citizen-science-enabled catalogue of the vaginal microbiome and associated factors. Nat. Microbiol. 8, 2183–2195 (2023).
Ahannach, S., Van Hoyweghen, I., Verhoeven, V. & Lebeer, S. Citizen science as an instrument for women’s health research. Nat. Med. 30, 3445–3454 (2024).
Condori, S. et al. Recent insights into the vaginal microbiota. Microb. Health Dis. 4, 1–12 (2022).
Ehlers, C., Ivens, U. I., Møller, M. L., Senderovitz, T. & Serup, J. Females have lower skin surface pH than men. Skin Res. Tech. 7, 90–94 (2001).
Luebberding, S., Krueger, N. & Kerscher, M. Skin physiology in men and women: In vivo evaluation of 300 people including TEWL, SC hydration, sebum content and skin surface pH. Int. J. Cosmet. Sci. 35, 477–483 (2013).
Giacomoni, P. U., Mammone, T. & Teri, M. Gender-linked differences in human skin. J. Dermatol. Sci. 55, 144–149 (2009).
Ying, S. et al. The influence of age and gender on skin-associated microbial communities in urban and rural human populations. PLoS One 10, e0141842 (2015).
Cauwenberghs, E. et al. Salivary microbiome of healthy women of reproductive age. mBio 14, https://doi.org/10.1128/mbio.00300-23 (2023).
Sirichoat, A. et al. Comparison of different hypervariable regions of 16S rRNA for taxonomic profiling of vaginal microbiota using next-generation sequencing. Arch. Microbiol. 203, 1159–1166 (2021).
Meisel, J. S. et al. Skin microbiome surveys are strongly influenced by experimental design. J. Invest. Dermatol. 136, 947 (2016).
L van der, M. aaten & Hinton, G. Visualizing Data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Perez, G. I. P. et al. Body site is a more determinant factor than human population diversity in the healthy skin microbiome. PLoS ONE 11, e0151990 (2016).
Shaw, J. A. et al. Oestrogen receptors alpha and beta differ in normal human breast and breast carcinomas. J. Pathol. 198, 450–457 (2002).
Howard, B. et al. Aging-associated changes in the adult human skin microbiome and the host factors that affect skin microbiome composition. J. Invest. Dermatol. 142, 1934–1946.e21 (2022).
Edmonds-Wilson, S. L., Nurinova, N. I., Zapka, C. A., Fierer, N. & Wilson, M. Review of human hand microbiome research. J. Dermatol. Sci. 80, 3–12 (2015).
Ross, A. A., Doxey, A. C. & Neufeld, J. D. The skin microbiome of cohabiting couples. mSystems 2, https://doi.org/10.1128/msystems.00043-17 (2017).
Ahannach, S. et al. Microbial and seminal traces of sexual intercourse and forensic implications. Res. Square https://doi.org/10.21203/RS.3.RS-4302243/V1 (2024).
Ghemrawi, M. et al. The genital microbiome and its potential for detecting sexual assault. Forensic. Sci. Int. Genet. 51, 102432 (2021).
Williams, D. W. & Gibson, G. Classification of individuals and the potential to detect sexual contact using the microbiome of the pubic region. Forensic. Sci. Int. Genet. 41, 177–187 (2019).
Wittouck, S., Rillaer, T., Van, Smets, W. & Lebeer, S. Tidytacos: An R package for analyses on taxonomic composition of microbial communities. J. Open Source Softw. 10, 6313 (2025).
Van Der Maaten, L., Courville, A., Fergus, R. & Manning, C. Accelerating t-SNE using tree-based algorithms. J. Mach. Learn. Res. 15, 3221–3245 (2014).
Acknowledgements
We extend our heartfelt thanks to all Isala volunteers and citizens who co-created this project with us. Their enthusiasm and commitment were the foundation of our work. We gratefully acknowledge the invaluable contributions of the following colleagues and students to the Isala sampling campaign and processing: I. Tuyaerts, N. Van Vliet, L. Van Ham, M. Legein, D. Vandenheuvel, A. Groenwals, S. El Messaoudi, T. Van Rillaer, J. Hiers, L. Van Dyck, C. Dricot, L. Leysen, and L. Martin Diaz. Strategic support was generously provided by L. Talboom and L. Haesevoets (Studio Maria, communication), C. Varszegi (Little Big Things, website: https://littlebigthings.be), the Antwerp Biobank (University Hospital Antwerp, Belgium; ID: BE 71030031000), and the Centre of Medical Genetics (University Hospital Antwerp, sequencing support). The authors acknowledge the European Research Council (starting grant Lacto-Be 852600 of S.L., with S.A., T.G., and S.W. appointed on the project), HORIZON EUROPE Framework programme (101213306 of S.L.), the Inter-University Special Research Fund of Flanders (iBOF; POSSIBL project supporting E.C. and BOF mandate FFB523 supporting M.H.), the industrial research fund UAntwerpen (IOF service platform microbiome sequencing), Flanders Innovation and entrepreneurship (HBC.2020.2873 supporting L.D.) and the Research Foundation—Flanders (FWO; post-doctoral research grant 12AZ624N of S.W., 12S4222N of I.D.B. and 1277222N of I.S., aspirant strategic basic research grant 1SD0622N of L.V.D. and Research projects G049022N, G031222N, S006424N of S.L.). The funders had no role in study design, data collection, data analysis, data interpretation or writing of the manuscript.
Author information
Authors and Affiliations
Contributions
S.A., G.D., V.V., E.O., S.W. and S.L. designed the study and worked on the conceptualization of the research project. S.A. and S.L. worked on the survey set-up and L.V.D., T.G., S.A. and I.S. cleaned the answers. S.A., L.V.D., E.C. and L.D. carried out the experimental and logistical work. I.D.B. and E.C. were responsible for the biobanking of all collected samples. T.G. and M.H. processed the sequencing data and performed the biostatistical analyses. L.V.D., T.G., S.V.B., M.H., S.A. and S.L. worked on the visualizations. L.V.D., T.G., S.A., S.V.D., M.H., C.N.A. and S.L. contributed to the interpretation of the results. L.V.D., S.A., T.G. and S.L. were responsible for the science communication to the participants in layman’s terms. S.L. was primarily responsible for funding acquisition and supervision of the study. L.V.D., S.A., T.G., S.V.B., M.H. and S.L. wrote the original manuscript. All authors contributed to reviewing and editing of the final manuscript.
Corresponding author
Ethics declarations
Competing interests
S.L. declares to be a voluntary academic board member of the International Scientific Association on Probiotics and Prebiotics (ISAPP, www.isappscience.org), cofounder of YUN and scientific advisor for Freya Biosciences. The team of S.L. declares research funding or consumables for research from YUN, BioOrg, Puratos, DSM-Firmenich, Drylocks Technologies and Lesaffre/Gnosis. G.D. is the chairperson of Femicare vzw (https://www.facebook.com/profile.php?id=100063440664962) and has worked as a medical consultant for various industries. None of these organizations or companies were involved in the design or data analysis of this study, which was fully funded by the university, governmental, and European funding. S.A. declares to be a voluntary member of the student and fellows association of ISAPP.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Vander Donck, L., Gehrmann, T., Ahannach, S. et al. The female intimate microbiome space. npj Womens Health 3, 55 (2025). https://doi.org/10.1038/s44294-025-00104-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s44294-025-00104-9