Abstract
Filoviruses such as Ebola virus are widely known as causative agents of severe human disease, although apathogenic filoviruses also exist. There is now increasing evidence that filoviruses circulate in almost all parts of the world, where they are being discovered in an expanding range of sometimes unexpected host species. Here we summarize the current knowledge regarding these novel filoviruses, and open questions that need answering to assess and prepare for the risk they pose.
Similar content being viewed by others
Classical filoviruses
Filoviruses (family Filoviridae) have long been known for their ability to cause severe disease in humans with extremely high case fatality rates1. The first member of this family to be recognized was Marburg virus (MARV), which was discovered in 1967 as a result of an outbreak of disease in laboratory workers in Germany and Serbia, who were involved in producing polio vaccine in kidney cells isolated from African Green monkeys (Chlorocebus aethiops) imported from Uganda2. Almost a decade later, in 1976, two related viruses, Ebola virus (EBOV) and Sudan virus (SUDV), were identified as the causative agents of parallel outbreaks of hemorrhagic fever in the Democratic Republic of Congo (DRC) and South Sudan, respectively3,4. While these viruses all resulted in a clinically very similar pattern of disease associated with hemorrhagic fever and multiorgan failure, they were found to be genetically distinct, with EBOV and SUDV forming the genus Orthoebolavirus, while MARV falls into the genus Orthomarburgvirus (Fig. 1). Since then several additional viruses have been identified as belonging to the Orthoebolavirus genus. These include Taï Forest virus (TAFV), which was identified after causing severe disease in a researcher working with nonhuman primates in the Taï Forest National park in Côte d’Ivoire in 1994, and represents the only confirmed case of TAFV infection to date5. More recently, Bundibugyo virus (BDBV) was discovered during a large hemorrhagic fever outbreak in Uganda in 20086. All of these human pathogenic viruses share overlapping endemic regions within central and western Africa (Fig. 2). They have also been shown, with greater or lesser degrees of evidence, to have bats as their natural reservoirs7. Consistent with this role as a reservoir host, infection of bats with these viruses does not appear to be associated with severe disease8,9. Interestingly, however, pigs have also been shown to be naturally susceptible to filoviruses found in both Africa and Asia, and can shed virus in the absence of overt disease, although there is currently no reported evidence for a role for transmission from pigs during outbreaks of human filovirus disease10,11,12,13,14,15.
A maximum-likelihood tree was constructed based on the filovirus polymerase (L) nucleotide sequences of ICTV-recognized species using data from the indicated GenBank accession IDs. Sequences were aligned using MUSCLE93 as implemented in MEGA1194. Trees were then inferred using the best-fit model (General Time Reversible; GTR). Results from 1000 bootstrap replicates are shown as percentages near the respective nodes. Mammalian filoviruses are indicated in green as circles, the sole Reptilian filovirus in red as a triangle, and fish filoviruses in blue as squares.
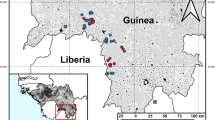
Locations from which novel filoviruses have been isolated (green stars) as well as locations where complete/coding-complete sequences (turquoise stars) or partial sequences (blue triangles) have been obtained are indicated. The arrow indicates the presumptive importation of diseased fish from Germany, into Switzerland, where the associated novel filovirus sequences were detected. Countries with serological evidence for novel filovirus circulation are indicated by light blue shading, whereas countries with reported cases of autochthonous filovirus disease are indicated by light green shading. BOMV Bombali virus, FIWIV Fiwi virus, HUJV Huángjiāo virus, LLOV Lloviu virus, LTBV Lötschberg virus, KNDV Kander virus, MLAV Měnglà virus, OBLV Oberland virus, TAPV Tapajós virus, XILV Xīlăng virus.
Despite the clear association of many filoviruses with human disease, apathogenic filoviruses have also long been known to exist, as exemplified by Reston virus (RESTV), which is also classified within the genus Orthoebolavirus, where it is most closely related to SUDV (Fig. 1). RESTV was first discovered in severely diseased nonhuman primate (rhesus macaques; Macaca mulatta) that were imported for research purposes from the Philippines into the US in 1989/9016, and subsequently also into Italy in 199217. No symptomatic human infections were reported in individuals with contact to these animals17,18, despite occupational exposure and/or seroconversion, suggesting that, in contrast to all other filoviruses known at that time, RESTV is apathogenic for humans. This conclusion was further supported by investigations of RESTV infections on pig farms in the Philippines in 2007/2008, with the outcome that more than 100 people have now been shown to have been infected with RESTV without any disease signs or symptoms18.
Evidence for (novel) filoviruses in geographically diverse regions
The decade after the discovery of BDBV saw an increasing number of filovirus outbreaks, including the dramatic EBOV epidemic in West Africa from 2014 to 2016, and a large and prolonged outbreak in the DRC from 2018 to 2020. Further, recent outbreaks of Marburg virus in Guinea, Ghana, Equatorial Guinea and Tanzania (reviewed in ref. 19), have dramatically expanded the known geographical range for not only MARV itself, but also for human pathogenic filoviruses, in general to the point where much of central and western Africa can now be considered to be endemic for one or more human pathogenic filoviruses (Fig. 2). Nonetheless, despite this increased awareness of the geographical extent of filovirus circulation within Africa, the diversity of known filoviruses remained restricted to the 6 classical viruses MARV, EBOV, SUDV, TAFV, RESTV, and BDBV. However, very recently, advances in high-throughput sequencing have resulted in a number of new filoviruses being identified, which have correspondingly resulted in a dramatic expansion of the filovirus family in recent years (Fig. 2) (Table 1). This has included the discovery of a novel member of the genus Orthoebolavirus, i.e. Bombali virus (BOMV), which was first identified in Angolan free-tailed bats (Mops condylurus) and little free-tailed bats (Chaerephon pumilus) in 2016 in Sierra Leone, and was subsequently also detected in M. condylurus in Kenya, Guinea, and Mozambique20,21,22,23,24. To date, human infections with BOMV have not been documented, despite one study examining patients with febrile illness in areas where BOMV has been found in bats24. It is also noteworthy that to date, despite its detection in several countries, isolation of BOMV from natural specimens has so far not been achieved, although a recombinant virus isolate has recently been generated25.
In addition to identifying novel orthoebolaviruses, virus discovery efforts have also revealed the existence of highly diverse filoviruses representing entirely novel genera. The first of these novel filoviruses was Lloviu virus (LLOV), which was assigned to the first new filovirus genus created since the 1970s, Cuevavirus (Fig. 1). LLOV RNA was first identified in samples from insectivorous bats (Schreiber’s long-fingered bats; Miniopterus schreibersii) taken in 2002 from a population roosting in a cave called Cueva del Lloviu in Spain during the investigation of a mass die-off in this bat species that was affecting populations in Spain, Portugal and France26 (Fig. 2). The virus was subsequently reported in the same bat species in Hungary in 2016, Italy in 2020, and Bosnia and Herzegovina in 202327,28,29. It is particularly worth noting that LLOV could be detected in the affected Hungarian bat colonies during surveillance from 2016 to 2020, and may have even been associated with an earlier die-off in 201328,30. Thus, these reports collectively suggest a widespread distribution and continual circulation of LLOV within continental Europe. Notably, a more recent study has also reported the successful isolation of LLOV from the blood of live-sampled Hungarian bats from the same region where the virus was previously detected30, strengthening the role of this species as a host for this virus (and perhaps as a reservoir species). Interestingly, urine and feces of LLOV-infected bats were negative for infectious virus in a recent study, and thus the mechanism of transmission between bats in the affected colonies remains open.
Identification of another novel bat filovirus, Měnglà virus (MLAV), was achieved following sequencing of RNA isolated from an unclassified Rousettus spp. bat from Měnglà County in Yunnan Province, China31 (Fig. 2). This virus is most closely related to Marburg virus, but also sufficiently distinct to warrant classification into a new genus, Dianlovirus (Fig. 1). However, to date, attempts to isolate infectious MLAV have been unsuccessful. Analysis of human serum samples from patients with febrile illness in the area surrounding where MLAV was detected showed that none contained antibodies against this virus; however, a caveat to this analysis is that positive bats were also not detected in the studied bat populations at that time32. Nonetheless, coupled with GPS tracking, which showed that the bats did not fly during the daytime (i.e. when human agricultural activities in the region are taking place) and rarely flew into residential areas32, these results have been taken to suggest that the risk of direct transmission of MLAV from infected bats to humans may be low under natural conditions.
Excitingly, these ever-accelerating virus discovery efforts have also revealed that filoviruses are not restricted to mammals, but rather that they can also be found in other animal classes, in particular in fish and reptiles. The first novel fish-associated filoviruses to be discovered were Xīlǎng virus (XILV; currently the only member of the genus Striavirus) and Huángjiāo virus (HUJV; the first member of the genus Thamnovirus), which were identified in striated frogfish and greenfin horse‐faced filefish, respectively, that had been captured in the East China Sea33 (Figs. 1 and 2). Since then a number of additional filoviruses have been found in diseased juvenile European perch imported into Switzerland from Germany34,35. Two of these viruses, i.e. Fiwi virus (FIWIV) and Kander virus (KNDV), group together with HUJV as part of the Thamnovirus genus, while the other two, Oberland virus (OBLV, genus Oblavirus) and Lötschberg virus (LTBV, genus Loebevirus) represent novel genera (Fig. 1). In addition to these fish-associated filoviruses, a novel reptile-associated filovirus, Tapajós virus (TAPV, genus Tapjovirus), was recently identified in RNA-seq data from a common lancehead viper sampled in Tapajós National Forest in Pará, Brazil36 (Figs. 1 and 2). Importantly, all of these novel filoviruses that have been detected in non-mammalian species are currently only known from their coding-complete genomes, with no infectious virus having been isolated, something that presents a major roadblock to their further study. In this regard it is also noteworthy that, while the genome of TAPV appears to closely resemble that of mammalian filoviruses, the genome organization of the fish-associated filoviruses is much more distinct in that it lacks homologs to some known filoviruses proteins, while also encoding unique putative open reading frames (reviewed in ref. 37).
In addition to these complete, or nearly complete, genome sequences, a wealth of partial sequences for other apparently novel (but as of yet unclassified) filoviruses have also been found in other fish and bat samples. These include the John dory filovirus and the Blue spotted goatfish filovirus, which were found in fish of the same name collected off the coast of New South Wales in Australia38, and a total of 24 filovirus sequences found mostly in Rousettus spp. and in a few cases in Eonycteris spp. bats in China and Vietnam39,40,41 (Fig. 2).
These genome-based studies indicating the widespread circulation of novel filoviruses are further corroborated by serological studies that have found antibodies against filoviruses, mainly in bats (Table 2), but sometimes also in humans and nonhuman primates (Table 3), in geographical regions that have to date not reported clinical filovirus infections (Fig. 2). Additionally, serological studies in endemic regions also indicate infection of other host species that have not previously been considered as targets of filovirus infection, such as pigs and dogs12,13,14,15,20,42,43,44,45,46. One of the first of these studies was conducted more than 30 years ago using human blood samples from healthy individuals in Germany, which showed a seroprevalence of 7% for filovirus antibodies by ELISA, of which ¾ of the positive results could be further confirmed by Western Blotting, prompting the authors to pose the question whether “subclinical filovirus infections occur worldwide?”47. While serological studies always carry some risk for false-positive findings, particularly if results are solely based on ELISA and not supported by confirmatory assays48, in their entirety, and particularly when taking into consideration the recent genomic data, it is now obvious that filoviruses are much more widespread than previously thought, and quite possibly occur worldwide, and that this occurs in many cases in the absence of an association with significant clinical disease.
Assessing the pathogenic potential of (novel) filoviruses
While there are continuous advancements being made in our understanding of the molecular biology of filoviruses (reviewed in ref. 49), virus discovery efforts and the appreciation that has accompanied them regarding the wide-spread distribution of both mammalian and non-mammalian filoviruses have only served to emphasize how much we still do not understand about the viral and host factors that determine the pathogenic potential of filoviruses, and consequently why some of these viruses are highly pathogenic (e.g. EBOV), whereas other are apathogenic in humans (e.g. RESTV). Nonetheless, also here progress is being made, and a number of factors have been suggested to play a role in this process, although the relevant factors also appear to differ between filoviruses. One of the earliest suggestions was that the filoviral glycoprotein plays a direct role in pathogenesis by causing cytotoxicity and increasing vascular permeability, as can be seen during expression of this protein in explanted human or porcine blood vessels50. However, experiments with chimeric viruses in which the glycoprotein genes were exchanged between EBOV and RESTV demonstrated that the glycoprotein, while contributing to the pathogenic potential of EBOV, is not sufficient to confer a highly pathogenic phenotype to RESTV51. This is also supported by work showing that expression of the EBOV glycoprotein is tightly controlled by several mechanisms during infection to prevent excess cytotoxicity, which further speaks against a decisive role of this protein in determining the pathogenic potential of filoviruses52.
Rather, a common feature that has been noted between RESTV, for which the available data strongly suggest that it is apathogenic in humans, and BOMV, for which infections in humanized mice indicate that this might also be the case25, is their reduced capacity for replication even in in vitro contexts. For RESTV this has been shown to be due to fundamental differences in the efficiency of viral RNA synthesis mediated by the viral replication machinery53,54. Intriguingly, although the underlying causes of attenuation of BOMV have not yet been well described, the limited available data rather indicate a defect related to the ability of the viral glycoprotein to mediate entry of viral particles25. Further, differences in the abilities of pathogenic and apathogenic filoviruses to modulate host antiviral responses to infection have been proposed to play a role in disease outcome. Indeed, a transcriptomics study comparing EBOV, RESTV and MARV indicated that the pathogenic EBOV and MARV were more capable of inhibiting interferon responses to infection55; however, such studies are inherently difficult due to the impaired growth of RESTV compared to these other classical filoviruses. Efforts to clarify this issue using in vitro reporter-based approaches independent of virus growth have also proven inconclusive, with one study of VP24, which is one of the two major viral type I interferon (IFN-I) antagonists, showing differences in the ability of EBOV and RESTV VP24 to impair the IFN-I response56. However, other studies did not observe such pathogenicity-related differences57,58,59,60. Similarly, for BOMV, one study found that the ability of its VP24 to inhibit the IFN-I response is impaired compared to both EBOV and RESTV59, while another study could not confirm these findings60. Ultimately though, in all these cases, experimental evidence that the observed differences are actually causative for the differences in pathogenic potential is still lacking.
Consequently, despite mounting molecular insights, we currently remain unable to predict the pathogenic potential of novel filoviruses based on their genotype, or specific viral traits, although poor in vitro growth (regardless of the molecular basis) appears to be highly suggestive. It is, therefore, currently necessary to evaluate the pathogenic potential of novel filoviruses in animal models. While the gold standard remains non-human primate models, it is clear based on infection with RESTV that these models can dramatically overestimate the lethality of apathogenic filoviruses (i.e. 50–83% lethality in cynomolgus macaques infected with RESTV, compared to 0% in humans)61,62. An alternative that has recently emerged are humanized mice63. Importantly, unlike many other small animal models, humanized mice do not require host adaptation of the virus, and reflect differences in pathogenic potential and virulence observed in humans, at least for orthoebolaviruses. Consequently, these animals were recently used to assess the pathogenic potential of a recombinant BOMV generated by reverse genetics (due to the absence of a natural isolate of this virus), with the results suggesting that BOMV resembles RESTV rather than EBOV with respect to its pathogenic potential in this model25. Similar results were later obtained using another animal model, i.e. IFN receptor-deficient mice, although here the authors noted limitations of this model with respect to its predictive value regarding the human pathogenic potential of filoviruses64.
Perspective
Our current lack of understanding regarding the underlying causes of differences observed in the pathogenic potential among filoviruses, together with concerns that mutation/evolution of apathogenic filoviruses might result in pathogenic phenotypes, is part of the reason why all mammalian filoviruses are currently routinely classified as biosafety level 4 (BSL4) pathogens. Importantly, this is regardless of whether or not they have a demonstrated potential to cause human disease, and in the case of RESTV despite significant evidence suggesting that human infections occur regularly but do not cause notable disease. The resulting need to work with any mammalian filovirus in maximum containment laboratories presents a significant obstacle to research on these viruses, and thus also to the development of countermeasures, and while this challenge is at least partially mitigated by the increasing availability of experimental systems that allow researchers to model the filovirus life cycle outside of such laboratories (reviewed in ref. 65), significant hurdles remain, particularly with respect to in vivo studies.
Importantly, this lack of detailed understanding regarding pathogenic determinants coupled with extremely divergent disease outcomes is something that is not specific for filoviruses, but rather it is common to all BSL4 virus families. Examples include the New World arenaviruses Junín and Machupo virus (highly pathogenic) and the related Tacaribe virus (apathogenic), the Old World arenavirus Lassa virus (highly pathogenic) and the related Mopeia and Mobala viruses (apathogenic), the orthonairovirus Crimean Congo Hemorrhagic Fever virus (highly pathogenic) and the related Hazara virus (apathogenic), and the henipaviruses Nipah and Hendra virus (highly pathogenic) and the related Cedar virus (apathogenic). Interestingly, unlike for filoviruses, in many countries these apathogenic viruses from other families are considered BSL3 or even BSL2 viruses (Table 4), and it is not clear what underlies these differences in risk perception for apathogenic filoviruses and for apathogenic relatives of other BSL4 viruses.
Given the much more wide-spread distribution of filoviruses that is being revealed by genomic and serological studies, we have to expect to increasingly detect these viruses also outside of areas that have been classically considered as filovirus endemic regions, and also not just in their natural reservoir hosts, i.e. bats, but in other susceptible species such as pigs or humans. However, the absence of reports of filovirus-like disease outside of these classical endemic regions, including in those with advanced diagnostic and medical infrastructure, clearly suggests that these filoviruses have a much lower pathogenic potential than classical filoviruses, and/or that there are barriers to their transmission to humans that are higher than in classical endemic regions. Importantly, the fact that serological studies indeed suggest exposure of humans to filoviruses outside of classical endemic regions, and also the fact that filoviruses as well as antibodies against such viruses have been detected in pigs, which have a much closer contact with humans than bats, speak towards the first possibility in at least some cases. Nevertheless, further research into filovirus transmission from bats to other hosts is clearly required to explain why filovirus outbreaks occur so rarely, or in some regions apparently not at all, despite their widespread distribution and continuous circulation. In addition, it will be paramount to better understand viral determinants of pathogenicity, in order to more accurately assess the risks posed by newly emerging filoviruses, and to prioritize resources and inform outbreak responses and public health measures. At the same time, a better understanding of the molecular basis for pathogenicity of known viruses could facilitate a rational reassessment of the possibility to perform research on apathogenic filoviruses outside of maximum containment laboratories. In particular for RESTV, where there is increasing evidence that it is apathogenic for humans, such a reassessment might be indicated.
Finally, given that there is an ever-increasing abundance of filoviruses being discovered, we also have to prepare for the emergence of a novel pathogenic filovirus species. In this context it will be important to be able to rapidly respond to such an occurrence, either by quickly adopting existing countermeasures, such as the existing vector vaccines that are now being used for the management of EBOV outbreaks66, or by developing broadly active therapeutics that not only target individual virus species, but rather are active against a wide range of viruses, including newly emerging (and potentially highly divergent) ones.
References
Izudi, J. & Bajunirwe, F. Case fatality rate for Ebola disease, 1976-2022: A meta-analysis of global data. J. Infect. Public Health 17, 25–34 (2024).
Slenczka, W. Filovirus Research: How it Began. Curr. Top. Microbiol. Immunol. 411, 3–21 (2017).
Pattyn, S., van der Groen, G., Jacob, W., Piot, P. & Courteille, G. Isolation of Marburg-like virus from a case of haemorrhagic fever in Zaire. Lancet 1, 573–574 (1977).
Bowen, E. T. et al. Viral haemorrhagic fever in southern Sudan and northern Zaire. Preliminary studies on the aetiological agent. Lancet 1, 571–573 (1977).
Le Guenno, B. et al. Isolation and partial characterisation of a new strain of Ebola virus. Lancet 345, 1271–1274 (1995).
Towner, J. S. et al. Newly discovered ebola virus associated with hemorrhagic fever outbreak in Uganda. PLoS Pathog. 4, e1000212 (2008).
Schuh, A. J., Amman, B. R. & Towner, J. S. Filoviruses and bats. Microbiol Aust. 38, 12–16 (2017).
Jones, M. E. et al. Experimental Inoculation of Egyptian Rousette Bats (Rousettus aegyptiacus) with Viruses of the Ebolavirus and Marburgvirus Genera. Viruses 7, 3420–3442 (2015).
Jones, M. E. B. et al. Clinical, Histopathologic, and Immunohistochemical Characterization of Experimental Marburg Virus Infection in A Natural Reservoir Host, the Egyptian Rousette Bat (Rousettus aegyptiacus). Viruses 11 https://doi.org/10.3390/v11030214 (2019).
Barrette, R. W. et al. Discovery of swine as a host for the Reston ebolavirus. Science 325, 204–206 (2009).
Marsh, G. A. et al. Ebola Reston virus infection of pigs: clinical significance and transmission potential. J. Infect. Dis. 204(Suppl 3), S804–S809 (2011).
Fischer, K. et al. Serological Evidence for the Circulation of Ebolaviruses in Pigs From Sierra Leone. J. Infect. Dis. 218, S305–S311 (2018).
Atherstone, C. et al. Investigation of Ebolavirus exposure in pigs presented for slaughter in Uganda. Transbound. Emerg. Dis. 68, 1521–1530 (2021).
Fischer, K. et al. Serological evidence of exposure to ebolaviruses in domestic pigs from Guinea. Transbound. Emerg. Dis. 67, 724–732 (2020).
Ogawa, H. et al. Detection of anti-ebolavirus antibodies in Ghanaian pigs. J. Vet. Med. Sci. 84, 1491–1494 (2022).
Jahrling, P. B. et al. Preliminary report: isolation of Ebola virus from monkeys imported to USA. Lancet 335, 502–505 (1990).
Viral haemorrhagic fever in imported monkeys. Wkly Epidemiol Rec. 67, 142–143 (1992).
Penas, J. A., Miranda, M. E., de Los Reyes, V. C., Sucaldito, M. N. L. & Magpantay, R. L. Risk assessment of Ebola Reston virus in humans in the Philippines. West. Pac. Surveill. Response J. 10, 1–8 (2019).
Marzi, A. & Feldmann, H. Marburg Virus Disease: Global Threat or Isolated Events? J. Infect. Dis. 228, 103–105 (2023).
Lebarbenchon, C. et al. Bombali Ebolavirus in Mops condylurus Bats (Molossidae), Mozambique. Emerg. Infect. Dis. 28, 2583–2585 (2022).
Goldstein, T. et al. The discovery of Bombali virus adds further support for bats as hosts of ebolaviruses. Nat. Microbiol. 3, 1084–1089 (2018).
Karan, L. S. et al. Bombali Virus in Mops condylurus Bats, Guinea. Emerg. Infect. Dis. 25, 1774–1775 (2019).
Kareinen, L. et al. Range Expansion of Bombali Virus in Mops condylurus Bats, Kenya, 2019. Emerg. Infect. Dis. 26, 3007–3010 (2020).
Forbes, K. M. et al. Bombali Virus in Mops condylurus Bat, Kenya. Emerg. Infect. Dis. 25, 955–957 (2019).
Bodmer, B. S. et al. In vivo characterization of the novel ebolavirus Bombali virus suggests a low pathogenic potential for humans. Emerg. Microbes Infect. 12, 2164216 (2023).
Negredo, A. et al. Discovery of an ebolavirus-like filovirus in europe. PLoS Pathog. 7, e1002304 (2011).
Toth, G. E. et al. Isolation and genome characterization of Lloviu virus from Italian Schreibers’s bats. Sci. Rep. 13, 11310 (2023).
Kemenesi, G. et al. Re-emergence of Lloviu virus in Miniopterus schreibersii bats, Hungary, 2016. Emerg. Microbes Infect. 7, 66 (2018).
Goletic, S. et al. Metagenomic Sequencing of Lloviu Virus from Dead Schreiber’s Bats in Bosnia and Herzegovina. Microorganisms 11 https://doi.org/10.3390/microorganisms11122892 (2023).
Kemenesi, G. et al. Isolation of infectious Lloviu virus from Schreiber’s bats in Hungary. Nat. Commun. 13, 1706 (2022).
Yang, X. L. et al. Characterization of a filovirus (Mengla virus) from Rousettus bats in China. Nat. Microbiol 4, 390–395 (2019).
Zhao, K. et al. Ecological study of cave nectar bats reveals low risk of direct transmission of bat viruses to humans. Zool. Res. 43, 514–522 (2022).
Shi, M. et al. The evolutionary history of vertebrate RNA viruses. Nature 556, 197–202 (2018).
Hierweger, M. M. et al. Novel Filoviruses, Hantavirus, and Rhabdovirus in Freshwater Fish, Switzerland, 2017. Emerg. Infect. Dis. 27, 3082–3091 (2021).
Seuberlich, T., Kuhn, J. H. & Schmidt-Posthaus, H. Near-Complete Genome Sequence of Lotschberg Virus (Mononegavirales: Filoviridae) Identified in European Perch (Perca fluviatilis Linnaeus, 1758). Microbiol. Resour. Announc 12, e0002823 (2023).
Horie, M. Identification of a novel filovirus in a common lancehead (Bothrops atrox (Linnaeus, 1758)). J. Vet. Med. Sci. 83, 1485–1488 (2021).
Hume, A. J. & Muhlberger, E. Distinct Genome Replication and Transcription Strategies within the Growing Filovirus Family. J. Mol. Biol. 431, 4290–4320 (2019).
Geoghegan, J. L. et al. Virome composition in marine fish revealed by meta-transcriptomics. Virus Evol. 7, veab005 (2021).
Yang, X. L. et al. Genetically Diverse Filoviruses in Rousettus and Eonycteris spp. Bats, China, 2009 and 2015. Emerg. Infect. Dis. 23, 482–486 (2017).
Makenov, M. T. et al. Detection of Filoviruses in Bats in Vietnam. Viruses 15 https://doi.org/10.3390/v15091785 (2023).
He, B. et al. Filovirus RNA in Fruit Bats, China. Emerg. Infect. Dis. 21, 1675–1677 (2015).
Sayama, Y. et al. A seroepidemiologic study of Reston ebolavirus in swine in the Philippines. BMC Vet. Res. 8, 82 (2012).
Pan, Y. et al. Reston virus in domestic pigs in China. Arch. Virol. 159, 1129–1132 (2014).
Haun, B. K. et al. Serological evidence of Ebola virus exposure in dogs from affected communities in Liberia: A preliminary report. PLoS Negl. Trop. Dis. 13, e0007614 (2019).
Fischer, K. et al. Ebola Virus Neutralizing Antibodies in Dogs from Sierra Leone, 2017. Emerg. Infect. Dis. 26, 760–763 (2020).
Allela, L. et al. Ebola virus antibody prevalence in dogs and human risk. Emerg. Infect. Dis. 11, 385–390 (2005).
Becker, S., Feldmann, H., Will, C. & Slenczka, W. Evidence for occurrence of filovirus antibodies in humans and imported monkeys: do subclinical filovirus infections occur worldwide? Med. Microbiol Immunol. 181, 43–55 (1992).
Formella, M. & Gatherer, D. The serology of Ebolavirus - a wider geographical range, a wider genus of viruses or a wider range of virulence? J. Gen. Virol. 97, 3120–3130 (2016).
Bodmer, B. S., Hoenen, T. & Wendt, L. Molecular insights into the Ebola virus life cycle. Nat. Microbiol. https://doi.org/10.1038/s41564-024-01703-z (2024).
Yang, Z. Y. et al. Identification of the Ebola virus glycoprotein as the main viral determinant of vascular cell cytotoxicity and injury. Nat. Med. 6, 886–889 (2000).
Groseth, A. et al. The Ebola virus glycoprotein contributes to but is not sufficient for virulence in vivo. PLoS Pathog. 8, e1002847 (2012).
Alazard-Dany, N. et al. Ebola virus glycoprotein GP is not cytotoxic when expressed constitutively at a moderate level. J. Gen. Virol. 87, 1247–1257 (2006).
Bodmer, B. S. et al. Differences in Viral RNA Synthesis but Not Budding or Entry Contribute to the In Vitro Attenuation of Reston Virus Compared to Ebola Virus. Microorganisms 8 https://doi.org/10.3390/microorganisms8081215 (2020).
Groseth, A., Feldmann, H., Theriault, S., Mehmetoglu, G. & Flick, R. RNA polymerase I-driven minigenome system for Ebola viruses. J. Virol. 79, 4425–4433 (2005).
Kash, J. C. et al. Global suppression of the host antiviral response by Ebola- and Marburgviruses: increased antagonism of the type I interferon response is associated with enhanced virulence. J. Virol. 80, 3009–3020 (2006).
Guito, J. C., Albarino, C. G., Chakrabarti, A. K. & Towner, J. S. Novel activities by ebolavirus and marburgvirus interferon antagonists revealed using a standardized in vitro reporter system. Virology 501, 147–165 (2017).
Schwarz, T. M. et al. VP24-Karyopherin Alpha Binding Affinities Differ between Ebolavirus Species, Influencing Interferon Inhibition and VP24 Stability. J. Virol. 91 https://doi.org/10.1128/JVI.01715-16 (2017).
Reid, S. P., Valmas, C., Martinez, O., Sanchez, F. M. & Basler, C. F. Ebola virus VP24 proteins inhibit the interaction of NPI-1 subfamily karyopherin alpha proteins with activated STAT1. J. Virol. 81, 13469–13477 (2007).
He, F. B. et al. Filovirus VP24 Proteins Differentially Regulate RIG-I and MDA5-Dependent Type I and III Interferon Promoter Activation. Front Immunol. 12, 694105 (2021).
Khan, H. et al. VP24 matrix proteins of eight filoviruses downregulate innate immune response by inhibiting the interferon-induced pathway. J. Gen. Virol. 104 https://doi.org/10.1099/jgv.0.001888 (2023).
Jahrling, P. B. et al. Experimental infection of cynomolgus macaques with Ebola-Reston filoviruses from the 1989-1990 U.S. epizootic. Arch. Virol. Suppl. 11, 115–134 (1996).
Fisher-Hoch, S. P. et al. Pathogenic potential of filoviruses: role of geographic origin of primate host and virus strain. J. Infect. Dis. 166, 753–763 (1992).
Escudero-Perez, B. et al. Comparative pathogenesis of Ebola virus and Reston virus infection in humanized mice. JCI Insight 4 https://doi.org/10.1172/jci.insight.126070 (2019).
Fletcher, P. et al. Pathogenicity of Lloviu and Bombali Viruses in Type I Interferon Receptor Knockout Mice. J. Infect. Dis. 228, S548–S553 (2023).
Wendt, L., Bostedt, L., Hoenen, T. & Groseth, A. High-throughput screening for negative-stranded hemorrhagic fever viruses using reverse genetics. Antivir. Res. 170, 104569 (2019).
Kallay, R. et al. Use of Ebola Vaccines - Worldwide, 2021-2023. MMWR Morb. Mortal. Wkly Rep. 73, 360–364 (2024).
Barr, J. et al. Detection of filovirus-reactive antibodies in Australian bat species. J. Gen. Virol. 103, https://doi.org/10.1099/jgv.0.001785 (2022).
Olival, K. J. et al. Ebola virus antibodies in fruit bats, bangladesh. Emerg. Infect. Dis. 19, 270–273 (2013).
Zhang, C. et al. Seroreactive Profiling of Filoviruses in Chinese Bats Reveals Extensive Infection of Diverse Viruses. J. Virol. 94, https://doi.org/10.1128/JVI.02042-19 (2020).
Yuan, J. et al. Serological evidence of ebolavirus infection in bats, China. Virol. J. 9, 236 (2012).
Dovih, P. et al. Filovirus-reactive antibodies in humans and bats in Northeast India imply zoonotic spillover. PLoS Negl. Trop. Dis. 13, e0007733 (2019).
Schulz, J. E. et al. Serological Evidence for Henipa-like and Filo-like Viruses in Trinidad Bats. J. Infect. Dis. 221, S375–S382 (2020).
De Nys, H. M. et al. Survey of Ebola Viruses in Frugivorous and Insectivorous Bats in Guinea, Cameroon, and the Democratic Republic of the Congo, 2015-2017. Emerg. Infect. Dis. 24, 2228–2240 (2018).
Djomsi, D. M. et al. Dynamics of Antibodies to Ebolaviruses in an Eidolon helvum Bat Colony in Cameroon. Viruses 14, https://doi.org/10.3390/v14030560 (2022).
Brook, C. E. et al. Disentangling serology to elucidate henipa- and filovirus transmission in Madagascar fruit bats. J. Anim. Ecol. 88, 1001–1016 (2019).
Ogawa, H. et al. Seroepidemiological Prevalence of Multiple Species of Filoviruses in Fruit Bats (Eidolon helvum) Migrating in Africa. J. Infect. Dis. 212(Suppl 2), S101–S108 (2015).
Ramirez de Arellano, E. et al. First Evidence of Antibodies Against Lloviu Virus in Schreiber’s Bent-Winged Insectivorous Bats Demonstrate a Wide Circulation of the Virus in Spain. Viruses 11, https://doi.org/10.3390/v11040360 (2019).
Nidom, C. A. et al. Serological evidence of Ebola virus infection in Indonesian orangutans. PLoS One 7, e40740 (2012).
Bouree, P. & Bergmann, J. F. Ebola virus infection in man: a serological and epidemiological survey in the Cameroons. Am. J. Trop. Med Hyg. 32, 1465–1466 (1983).
Gonzalez, J. P. et al. Antibody prevalence against haemorrhagic fever viruses in randomized representative Central African populations. Res Virol. 140, 319–331 (1989).
Paix, M. A. et al. [Serological study of the virus responsible for hemorrhagic fever in an urban population of Cameroon]. Bull. Soc. Pathol. Exot. Filiales 81, 679–682 (1988).
Saluzzo, J. F., Gonzalez, J. P., Herve, J. P., Georges, A. J. & Johnson, K. M. [Preliminary note on the presence of antibodies to Ebola virus in the human population in the eastern part of the Central African Republic]. Bull. Soc. Pathol. Exot. Filiales 73, 238–241 (1980).
Meunier, D. M. et al. [Current serologic data on viral hemorrhagic fevers in the Central African Republic]. Bull. Soc. Pathol. Exot. Filiales 80, 51–61 (1987).
Johnson, E. D., Gonzalez, J. P. & Georges, A. Filovirus activity among selected ethnic groups inhabiting the tropical forest of equatorial Africa. Trans. R. Soc. Trop. Med Hyg. 87, 536–538 (1993).
Johnson, E. D., Gonzalez, J. P. & Georges, A. Haemorrhagic fever virus activity in equatorial Africa: distribution and prevalence of filovirus reactive antibody in the Central African Republic. Trans. R. Soc. Trop. Med Hyg. 87, 530–535 (1993).
Gonzalez, J. P., Nakoune, E., Slenczka, W., Vidal, P. & Morvan, J. M. Ebola and Marburg virus antibody prevalence in selected populations of the Central African Republic. Microbes Infect. 2, 39–44 (2000).
Nakounne, E., Selekon, B. & Morvan, J. [Microbiological surveillance: viral hemorrhagic fever in Central African Republic: current serological data in man]. Bull. Soc. Pathol. Exot. 93, 340–347 (2000).
Tignor, G. H., Casals, J. & Shope, R. E. The yellow fever epidemic in Ethiopia, 1961-1962: retrospective serological evidence for concomitant Ebola or Ebola-like virus infection. Trans. R. Soc. Trop. Med Hyg. 87, 162 (1993).
Mathiot, C. C., Fontenille, D., Georges, A. J. & Coulanges, P. Antibodies to haemorrhagic fever viruses in Madagascar populations. Trans. R. Soc. Trop. Med Hyg. 83, 407–409 (1989).
Tomori, O., Fabiyi, A., Sorungbe, A., Smith, A. & McCormick, J. B. Viral hemorrhagic fever antibodies in Nigerian populations. Am. J. Trop. Med Hyg. 38, 407–410 (1988).
Changula, K. et al. Serological Evidence of Filovirus Infection in Nonhuman Primates in Zambia. Viruses 13 (2021). https://doi.org/10.3390/v13071283
Blackburn, N. K., Searle, L. & Taylor, P. Viral haemorrhagic fever antibodies in Zimbabwe schoolchildren. Trans. R. Soc. Trop. Med Hyg. 76, 803–805 (1982).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32, 1792–1797 (2004).
Tamura, K., Stecher, G. & Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 38, 3022–3027 (2021).
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Groseth, A., Hoenen, T. Novel filoviruses: indication of a global threat or cause to reassess our risk perception?. npj Viruses 2, 38 (2024). https://doi.org/10.1038/s44298-024-00050-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s44298-024-00050-4
This article is cited by
-
In-silico evaluation of Azadirachta indica-derived Daucosterol against key viral proteins of Ebolavirus using ML and MD simulations approach
Journal of Biological Physics (2025)