Abstract
Dietary composition can significantly influence health and lifespan, however, robust knowledge on which food components, at what concentration exert which long-term health effects is still incomplete. Here, we explored the effects of dietary protein intake on Ercc1Δ/− DNA-repair-deficient mice, which are an excellent model for accelerated ageing and are hyperresponsive to the anti-ageing effect of dietary restriction. Restricting dietary protein by 50% extended lifespan in male mice, but not in females. Restricting protein levels beyond 80% improved various neurological health parameters, while a further reduction to 95% affected appetite and became distinctly detrimental. Conversely, a near doubling of protein intake and isocaloric compensatory lowering with carbohydrates significantly shortened lifespan in both sexes. Gene expression analysis of liver from mice on a high-protein, low-carbohydrate diet to those on high-carbohydrate, low-protein revealed increased expression of oxidative phosphorylation, enrichment of processes associated with tissue injury, inflammation, and gene-length-dependent transcriptional decline (GLTD), recently shown to reflect DNA damage accumulation causing transcription stress, and cellular ageing. Finally, GLTD was also identified by reanalysis of publicly available data of wild-type mice, rats and humans on high-protein diets, suggesting that increased dietary protein enhances GLTD and accelerates systemic ageing. Together, our findings have implications for nutritional guidelines for progeroid DNA-repair-deficient human syndromes, warrant the use of excessive protein intake for sustaining health, and suggests GLTD as a sensitive read-out of overall health and predictor of biological ageing.

Similar content being viewed by others
Introduction
Life expectancy worldwide has doubled in the last century, with projections indicating that the number of people older than 80 will triple between 2020 and 20501. Many of these additional years are, however, spent with disabilities and disease. This development presents substantial economic, societal and healthcare challenges, necessitating a deeper understanding of ageing and means to ameliorate it2,3. Ageing can be characterized by a variety of hallmarks, including genomic instability, loss of proteostasis, and inflammaging4,5, with accumulating DNA damage recently highlighted as root cause6,7,8. The consequent dysregulation of numerous cellular processes affects health, apparent from compromised metabolic functions, diminished regenerative capabilities, and increased disease vulnerability9,10. Interestingly, modulating metabolic processes e.g., through dietary changes and/or exercise, has been shown to improve both health parameters and lifespan11,12,13,14.
While diets are commonly associated with weight management, their broader impact is evident in therapeutic applications, such as ketogenic and time-restricted feeding diets to counteract obesity, type 2 diabetes, and neurological events such as seizures15,16,17,18. However, dietary restriction (DR), defined as a reduction of all dietary components without malnutrition, remains the most extensively studied intervention in the context of ageing19. DR is associated with numerous health benefits and lifespan extension across species ranging from yeast to monkeys20. Furthermore, 12% DR in humans improves cardiac, metabolic, and inflammatory markers, although adhering to a DR lifestyle can be challenging21,22.
We previously showed a remarkable lifespan extension by DR in progeroid Ercc1Δ/− DNA repair-deficient mice with prominent health benefits noted across all organs23,24. These mice, carrying both a null and a 7 amino acid C-terminally truncated Ercc1 allele, exhibit only 5–10% of active ERCC1 protein25. This deficiency impairs the ERCC1-XPF complex, which acts as pivotal endonuclease in various DNA repair processes, including Global Genome Nucleotide Excision Repair (GG-NER), Transcription-Coupled Repair (TCR), interstrand cross-link repair, and single-strand annealing repair of double-strand breaks26. Consequently, a broad range of DNA lesions persists and accumulates with age, leading to rapid ageing and a lifespan of 4–6 month on a standard ad libitum diet23,27. Within this period, these mice exhibit numerous systemic age-related phenotypes, such as dramatic neurodegeneration, tremors, imbalance, and frailty, which were all drastically delayed by DR28,29,30,31. As these mice turned out to be highly sensitive to nutritional interventions23,24, we here explored the effect of dietary protein intake in relation to lifespan and health, and found a potential link with DNA damage.
Results
Protein restriction slightly extends lifespan in Ercc1 Δ/- mice
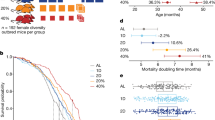
Modification of dietary protein, mostly restriction, has previously been shown to influence health and lifespan in multiple species32,33,34,35. To examine the effect of dietary protein restriction (PR) on ageing, we subjected progeroid Ercc1Δ/- mice to ad libitum (AL) fed diets reduced in protein by 30, 50, or 80 percent. The diets were supplemented with dextrose to improve palatability (See methods for more information). All diets were based on AIN93G (%Kcal: 19.3 protein, 64.0 carb, 16.7 fat; caloric density 3.97 Kcal/gm food) which served as AL-control. There was no difference in food intake nor bodyweight between diets (Fig. 1A, B, Supplementary Fig. 1A–F), although we noted a uniform increase compared to previous Ercc1Δ/- cohorts in which no dextrose supplementation was employed23,27,36. Specifically in males did we observe a minor but significant increase in median lifespan from 18.4 to 21.9 weeks ( ~ 3.5 weeks, +19%, p = 0.008) only by 50% PR (Fig. 1C). No significant lifespan changes were noted for female Ercc1Δ/- mice on these diets (Fig. 1D). As benefits for mice have previously been shown with protein restriction levels beyond 80%37, we next expanded the range of PR and tested reductions between 80 and 95 percent, and this time excluded dextrose from the diets to avoid an enhanced food intake. Here, we noted a gradual decrease in food intake with lower protein content (Supplementary Fig. 1G, H), which in turn correlated with lower body weights (Fig. 1E, Supplementary Fig. 1I). Moreover, increased levels of protein restriction were associated with a delayed onset of neurological phenotypes, such as tremors and imbalance, with the delay being most notable at 93% PR (Fig. 1F, G). Concomitantly, 93% PR increased median lifespan from 19.5 to 22.5 weeks ( ~ 3 weeks, +15%, p = 0.003), while a further reduction to 95% still delayed neurological phenotypes but became distinctly detrimental for lifespan ( ~ 2.4 weeks, -12%, p = 0.0102) (Fig. 1H, Supplementary Fig. 1J), consistently found for males and females (Supplementary Fig. 1K–N). In conclusion, while protein restriction up to 93% seemed to be associated with a slight extension of several health parameters and lifespan, this effect may also at least in part be explained by animals imposing dietary restriction upon themselves.
A, B Body weight trajectories of Ercc1Δ/- mice across different protein restriction (PR) diets (30%, 50%, and 80%) in both male (A) and female (B) cohorts. C, D Survival curves of the same groups for male (C) and female (D) mice. E Body weight trajectories of mice subjected to either control or 80–95% PR diets. F, G Graphs depicting the phenotypical onset of tremors (F) and imbalance (G). H Lifespan curves for the same dietary regimens. Data are presented as mean ( ± SD). Black arrows indicate when diets were started. Significance levels are denoted as *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 and compared to the respective control group.
Increased dietary protein-to-carbohydrate content reduces Ercc1 Δ/- lifespan
Conversely, diets high in protein have been observed to impact long-term health in young and middle-aged organisms34,38,39. Therefore, we next modified macronutrient ratios in the diets of Ercc1∆/− mice, reducing one macronutrient by 30% and isocalorically compensating with another. While dietary reference values are higher for carbohydrate intake compared to fat and protein, low carbohydrate diets are generally associated with various health benefits in humans13,40,41. Therefore, we compensated diets reduced in fat or protein with carbohydrate and, as proteins and carbohydrates have the same caloric content, we compensated the reduced carbohydrate diet with protein. Diets were abbreviated as LFHC (Low-Fat/High-Carbohydrate), LCHP (Low-Carbohydrate/High-Protein), and LPHC (Low-Protein/High-Carbohydrate) (Fig. 2A, Supplementary Fig. 2A). DR and low protein were only tested for one gender, namely in females and males respectively. Neither LFHC nor LPHC affected food consumption or body weight (Fig. 2B, C, Supplementary Fig. 2B–G). Lifespan analysis also revealed no significant changes with LFHC and LPHC diets (Fig. 2D–F). However, while both 30% DR and LCHP diet resulted in lower body weight due to lower food intake only DR drastically extended lifespan (Fig. 2B–F), consistent with prior studies23. In contrast, LCHP significantly shortened median lifespan from 16.0 to 13.1 ( ~ 3 weeks, -18%, p = 3.04E-05) in males and from 20.7 to 13.30 ( ~ 7.5 weeks, -36%, p = 2E-06) in females, as well as maximal lifespan in both sexes (Fig. 2D, E). Combined, these results indicate that while severely lowered protein diets may confer modest benefits, high protein, low carbohydrate diet significantly shortens the lifespan of Ercc1Δ/- mice in both sexes (Fig. 2F).
A Simplified table of isocalorically maintained macro nutritional changes in low-fat, high-carbohydrate (LFHC), low-carbohydrate, high-protein (LCHP), and low-protein, high-carbohydrate (LPHC) diets relative to AIN93G control (caloric density 3.97 Kcal/gm food). B, C Trajectories of body weight changes in male (B) and female (C) mice with data presented as mean ( ± SD). D, E Kaplan-Meier curves depicting survival of male (D) and female (E) mice. F Forest plot depicting log10 hazard ratios for different dietary interventions with 95% confidence intervals as indicated. Black arrows indicate when diets were started. Significance is denoted as ****p ≤ 0.0001 and compared to the respective control group.
Increased dietary protein-to-carbohydrate content alters metabolism and tissue-damage-related pathways
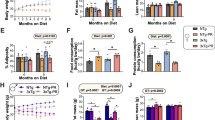
To further explore the impact of macronutrient modifications on lifespan and parameters of ageing, particularly the relationship between high and low protein intake in relation to carbohydrates, we analysed the transcriptional profiles of liver from 11-week-old male Ercc1∆/− mice, maintained for about 3–4 weeks on the diets described above. At 11 weeks of age, the mice are still relatively healthy and do not exhibit severe ageing pathologies. The liver was chosen as the primary organ handling nutritional intake, including detoxification and metabolic response, while later in life displaying dramatic features of accelerated ageing23,27,42. Principal component analysis (PCA) revealed a high degree of data homogeneity hampering group separation, which may be in line with the relatively brief period of dietary changes (Supplementary Fig. 3A). Therefore, we focused on the samples with the strongest contrasts in terms of biological outcome: comparing LCHP and LPHC diets. Gene-set enrichment analysis (GSEA) revealed metabolic alterations characterized by increased oxidative phosphorylation, and upregulation of Irs1 and Irs2 gene targets, alongside downregulation of Srebf targets, suggesting enhanced aerobic energy metabolism on the LCHP diet (Fig. 3A–C, Supplementary File 1). GSEA furthermore signalled reduced peroxisome activity in LCHP vs LPHC, indicative of altered fatty acid metabolism and reactive oxygen species handling (Supplementary Fig. 3B). Additionally, we observed a subtle trend towards increased cellular stress characterized by Tgfb1, inflammatory pathways, and Foxo3 signalling (Fig. 3D, E, Supplementary Fig. 3C). This was further supported by Ingenuity Pathway Analysis (IPA) of the 307 differentially expressed genes (DEG, FDR < 0.05) between LCHP and LPHC, which indicated significant enrichment of regulatory cytokines, including IL1β, IL4, IL6, LIF, and IL13 (Fig. 3F), many of which have been associated with impaired liver functioning like steatosis and fibrosis and consistent with elevated cellular senescence and inflammation43,44,45. Analysis of key signalling pathways and liver-specific toxicity using IPA similarly indicated an enrichment of processes related to liver dysfunction (Fig. 3G, Supplementary Fig. 3D). Altogether, gene expression provided hints towards increased toxicity by the LCHP diet, resembling further accelerated ageing.
A–C GSEA plots based on male liver tissues (n = 4 per group) comparing the effect of low-carbohydrate, high-protein (LCHP), and low-protein, high-carbohydrate (LPHC) diets shown for oxidative phosphorylation (A), targets of Irs1 and Irs2 (B), and Srebf targets (C). D, E GSEA plots indicating the expression profile of Tgfb1 targets (D) and LPS induced inflammatory response signalling (E). F, G Bar graphs depicting IPA based top significantly enriched upstream regulatory cytokine expression (F), and signalling pathways filtered for z-score > |0.5| (G). H Graphical representation of gene-length-dependent transcriptional decline (GLTD), ordered by gene size and categorized in bins of 500 (N = 13.426). I GSEA figure depicting the expression of the Tp53-regulated metabolic gene pathways (J) Top statistically significant pathways identified by GSEA analysis from the Tabula Muris Senis ageing mouse database. Diets are indicated as LFHC (low-fat, high-carbohydrate), LCHP (low-carbohydrate, high-protein), and LPHC (low-protein, high-carbohydrate). In the graphs, red indicates upregulation with age, while blue denotes downregulation with age, dot size denoted size of the gene set.
Increased dietary protein-to-carbohydrate content exacerbates transcription stress and ageing in Ercc1 Δ/- mice
DNA damage is central to the process of systemic ageing7, and is among others associated with sterile inflammation and tissue damage46,47. Recent studies have further shown that ageing is accompanied by gene-length-dependent transcription decline (GLTD), also known as transcription stress, linked to the accumulation of RNA-polymerase-stalling stochastic DNA damage8. The transcriptional decline of long genes with age was initially discovered by us in Ercc1Δ/- mice, wild-type rat livers, and the ageing human hippocampus23, and has since been confirmed in a wide range of organisms from worms to human, tissue types, as well as in individuals with neurodegenerative disease48,49,50, indicating its universal occurrence and strong evolutionary conservation and consistent with the notion that accelerated ageing of the Ercc1 mutant is highly relevant for normal ageing.
Here, we sorted all genes expressed in the liver (N = 13.426) based on gene length and clustered them in bins of 500, revealing that increased dietary protein intake triggers GLTD as indicated by a statistically highly significant downregulation of longer genes in LCHP vs LPHC diets (Fig. 3H, p = 1.89E-12). Additional assessment using GSEA suggested increased TP53-mediated regulation of metabolic genes, genotoxic damage, RNA metabolism, and ageing (Fig. 3I, Supplementary Fig. 3E–I), in line with the presence of GLTD. Furthermore, using cell type-specific signature gene sets from the Molecular Signatures Database (MSigDB) in GSEA, we identified nine enriched signatures from the Tabula Muris Senis ageing dataset. This data consisted of cell type-specific expression changes between old and young mice. Importantly, all nine, except one small gene set ‘kidney T cell ageing’, were positively enriched in our dataset concordant with ageing (Fig. 3J), consistent with the idea that shifting dietary content from carbohydrates towards an increased intake of dietary protein accelerates ageing. As the above transcriptomic changes could have been confounded by the effects of the LPHC diet used as control, we repeated our analysis comparing LCHP vs AL-control. Herein, again increased oxidative phosphorylation by GSEA, enrichment of regulatory cytokines by IPA and GLTD-related transcription stress were observed by the LCHP diet although less pronounced (Supplementary Fig. 4A–C).
Increased dietary protein exacerbates transcriptional parameters of GLTD and ageing also in conditions of proficient DNA repair
To further explore the hypothesis that an increased protein-to-carbohydrate ratio leads to elevated GLTD, we explored all currently available RNA expression datasets that investigated elevated protein intake in combination with health parameters. Applying a diet of 30% kcal from protein versus 5% kcal for 12 weeks, likewise compensated with carbohydrates, to C57BL/6 J wild-type mice did not affect energy intake or adiposity51. However, assessing their transcriptome data of hypothalamus we observed for expressed genes (N = 11.500), as well as DEGs (N = 829), between LCHP and LPHC (with protein/carb/fat ratios of 30/50/20 and 05/75/20 respectively), a clear gene-length-dependent decline in transcriptional output, associated with a higher protein intake (Fig. 4A, B, p = 4.96E-13 and p < 1E-13 respectively), similar to our own liver dataset and indicative of elevated damage. Additionally, GSEA showed that 88 out of 90 significantly enriched Tabula Muris Senis Ageing signatures were increased (Fig. 4C, Supplementary File 2), supporting the idea of accelerated ageing.
A Graphical representation of gene-length-dependent transcriptional decline (GLTD) in hypothalami, ordered by gene size and categorized bins of 500 from high vs low protein diets (N = 11.500). B Boxplots based on gene length of up/down regulated DEGs of the same comparison. C Top 30 most statistically significant pathways identified by GSEA analysis from the Tabula Muris Senis database of the same comparison. High protein (HP), low protein (LP) (D, E) Boxplots based on gene length of up/down regulated DEGs from liver in mice fed 60% vs 5% (D) or 42% vs 14% (E) protein. F, G Boxplots based on gene length of up/down regulated DEGs due to an altered macro nutritional balance from rat liver (F) and human subcutaneous adipose tissue (G). In the graphs, red indicates upregulation, while blue denotes downregulation. Significance levels are denoted as ‘ns’ for p > 0.05 and ****p ≤ 0.0001.
Furthermore, re-analysis of DEGs from micro-arrays of livers of 15-month-old C57BL/6 J wild-type mice maintained for 14 months on diets with protein/carb/fat ratios of 60/20/20 versus 05/75/20 and 42/29/29 versus 14/57/2952, again confirmed the selective suppression of longer genes by a higher protein lower carbohydrate intake (Fig. 4D, E). Interestingly, the same authors previously showed increased lifespan and metabolic health with LPHC diets and reduced lifespan and metabolic health with LCHP diets34. The data presented here using the progeroid Ercc1 repair mutant are in line with these findings in wild-type mice and further strengthen the parallels and validity of the accelerated ageing model for assessing the effect of nutritional interventions on normal ageing. Additionally, we re-analysed livers from 6-month-old wild-type male Wistar rats fed a high protein (45/45/10) or standard control protein diet (20/70/10) for 4 months53, and human subcutaneous adipose tissue biopsies from overweight middle-aged subjects adhering to a 25% energy-restricted diet, either combined with high protein (1.5 g/kg-bw/d) or standard protein (0.8 g/kg-bw/d) for 12 weeks54. Although these datasets consisted of low number of DEGs, both showed a non-significant trend in downregulation of longer genes (Fig. 4F, G), thus potentially extending our findings of GLTD induced by high protein diets to other organs and species.
Discussion
Dietary modifications can have a major impact on both health and lifespan, with the most well-studied intervention being dietary restriction (DR). However, due to practical challenges of sustaining long-term DR, it is important to explore alternative dietary strategies. Here, we focused on varying protein levels on the health and lifespan of DNA-repair-deficient Ercc1Δ/- mice, which display numerous features of accelerated but bona fide ageing and respond exceptionally well to DR23,27,28. We found that protein restriction (PR) enhanced health and lifespan to some extent, with mildly more pronounced effects in males—a finding that largely mirrors observations in wild-type animals32,55,56,57. Although only observed with limited animal numbers used in this study, diets in which protein is reduced by ≥80% led to a reduced food intake, despite these diets typically being associated with increased consumption due to the satiating effects of dietary protein. This reduction could have been caused by altered taste and the absence of additional dextrose in more stringent PR diets, likely altering their palatability. Despite the reduced food intake with very low dietary protein, the health and lifespan benefits observed were much smaller than those seen with DR23(and Fig. 2E). This discrepancy may stem from excessively low protein levels, impairing optimal metabolism. Alternatively, the difference in feeding behaviour induced by DR and PR might also have contributed. While DR is usually provided as a fixed amount once per day, which promotes rapid consumption of food, with subsequent fasting, mice on PR diets consumed smaller quantities over extended periods, alike ad libitum, limiting any potential fasting benefits58. However, mechanistically both PR and DR have been shown in part to activate stress resistance genes via attenuation of GCN2, mTOR, and GH/IGF-1 nutrient sensing pathways and modulate metabolism via altering acetyl-CoA production59.
On the other hand, low carbohydrate high dietary protein was associated with a remarkable shortened health- and lifespan. The LCHP diet in fact reduced food intake comparable to 30% DR, while only DR dramatically improves health and extends life in DNA-repair-deficiency mutants23. The reduced food intake was likely caused by the appetite-suppressing effects of increased dietary protein60, but without yielding benefits alike DR. We attribute the negative effects to the high protein as low carbohydrates are generally considered beneficial13,40,41. To obtain clues on underlying mechanisms we analysed gene expression changes in liver contrasting LPHC with LCHP. While datasets demonstrated a strong GLTD effect, the significance of DNA-damage-related processes was low, suggesting that external confounding factors cannot be ruled out. These could include immune system adaptations, changes in cell type distribution, increased expression of short stress-responsive genes, or alterations in RNA metabolism48,61. Interestingly, both our own liver RNA sequencing data and the deposited mouse hypothalami dataset (GSE114960), showed a highly significant enrichment of Muris Tabula Senis Ageing profiles. This effect was even more pronounced in the wild-type dataset, potentially due the longer high-protein feeding regimen. The parallel between our high-protein liver expression profile and ageing may make sense in the light of our finding that the dataset in both cases is dominated by GLTD.
Mechanistically, increased dietary protein upregulated genes of the oxidative phosphorylation pathway, an effect likely driven by an altered protein-to-carbohydrate ratio, impacting both energy metabolism and anabolic processes55,62. This metabolic shift, combined with elevated ferroptosis signalling and production of nitric oxide and reactive oxygen species, indicates disrupted reactive oxygen homeostasis, likely leading to increased overall macromolecular damage, inflammation and the observed tissue damage and fibrosis63,64,65. However, it remains to be investigated what amount and types of reactive metabolites are more produced upon increased metabolism by a high protein intake. Noteworthy are aldehydes (e.g. produced from catabolism) which were recently shown to exacerbate the effects of NER and Fanconi Anemia DNA-repair-deficiency in mice66,67. This hypothesis of increased reactive metabolite production by high protein diets is in line with the observed strongly increased GLTD, which we found in normal ageing to be driven by accumulating stochastic DNA damage, interfering with transcription and we found to be associated with ageing8,23. The link between increased protein consumption, health, and lifespan may reflect an evolutionary trade-off, optimizing early life fitness and growth at the expense of longevity68. Surprisingly, several genes of the GCN2, mTOR, and GH/IGF-1 nutrient sensing pathways are relatively long, making them more susceptible to GLTD, counteracting any presumed stimulation by increased amino acids. Interestingly, these ageing-related effects are likely independent of mTOR signalling, which is typically increased by high-protein diets, as its inhibition by Rapamycin is known not to impact the lifespan of Ercc1Δ/− mice36.
Our findings, while based on rodent models, highlight the need for a more comprehensive evaluation of high-protein, low-carbohydrate diets. These diets are widely used for weight loss or build-up of muscle mass in humans but also associated with increased risk of disease and mortality38,39,69,70,71, potentially mediated by processes observed in our study. In contrast, low dietary protein intake is linked with improved health in individuals under 65 while such benefits are not observed in older adults, suggesting age-related metabolic changes that further complicate the underlying connections72. In fact, elderly people are commonly given food richer in proteins and fats to counteract muscle loss and weight loss. Our results rather argue against the use of excessive protein intake. As our repair-deficient mice constitute valid models for progeroid DNA repair deficiencies, such as Cockayne syndrome and Trichothiodystrophy our findings may also speak to nutritional guidelines for these rare disorders. To obtain a more comprehensive understanding future studies would benefit from including mice at different ages, increasing group sizes and extending the period of exposure to high-protein diets to improve assessment of long-term effects and potential health risks. It would be particularly interesting to include analysis of GLTD in these and human studies with dietary protein alterations with and without exercise. Furthermore, limiting confounding factors such as changes in food intake is crucial to accurately interpret the impact of high-protein diets. Finally, protein sources with different bioavailability (animal vs. plant) differentially impact health and ageing73,74,75. Investigating the molecular mechanisms behind these effects could provide essential insights not only aiding the refinement of dietary guidelines but also enhance our understanding of the interaction between diet, the microbiome, genetics, and ageing.
It is noteworthy, that our GLTD analysis was able to detect highly significant gene-expression changes already after four weeks of LCHP diet, when PCA did not reveal clear differences in overall gene expression. This indicates that GLTD may constitute a very sensitive read-out of genomic health, predicting later health effects and ageing trajectories. As future outlook, we intend to further explore GLTD as a valuable monitor of genomic health, which can inform about long-term beneficial or detrimental lifestyle factors, most notably nutrition, medical treatments and exposure to unknown compounds.
Methods
Ethics statement
The usage of animals in the experimental procedures was in accordance with the Principles of Laboratory Animal Care and with the guidelines approved by the Animal Ethical Committee of the National Institute for Public Health and the Environment (DEC no. 201100285, and 1200163) or Erasmus MC (DEC no. 139-12-12 and 139-12-13) in full accordance with European legislation (Recommendation 2007/526/EC).
Mouse model
The generation and characterization of Ercc1∆/+ and Ercc1+/− mice have been previously described23,27,76. Ercc1∆/− mice were obtained by crossing Ercc1∆/+ (in a pure C57BL6J or FVB background) with Ercc1+/− mice (in a pure FVB or C57BL6J background respectively) to yield Ercc1∆/− offspring with a genetically uniform F1 C57BL6J/FVB hybrid background. Typical unfavourable characteristics, such as blindness in an FVB background or deafness in a C57BL6J background, do not occur in this hybrid background. Wild-type F1 littermates or Ercc1∆/− mice on AIN93G control food were used as controls. Hence, all animals used in the studies described here were of the same healthier F1 C57BL6J/FVB hybrid background. Mice were clinically diagnosed daily in a blinded manner and were weekly weighed and visually inspected. Also, mice were scored in a for the technicians and animal care takers blinded fashion for gross morphological and motor abnormalities in experiments where so reported. All efforts were made to ameliorate the suffering of the animals.
Housing conditions
Animals were maintained in a controlled environment (20–22°C, 12 h light/dark cycle) and were housed in individual ventilated cages under specific pathogen-free conditions. Animals were group housed at the RIVM location (experiments with more than 10) when possible and individually housed at the EMC (experiments used less than 10 mice) location. Since the Ercc1Δ/− mice are smaller, food was administered within the cages and water bottles with long nozzles were used from around two weeks of age. Food and water were offered ad libitum (AL) unless stated otherwise.
Dietary interventions
All animals were bred and maintained on AIN93G synthetic pellets (Research Diet Services B.V., Wijk bij Duurstede, Netherlands; gross energy content 4.9 kcal/g dry mass, digestible energy 3.97 kcal/g, using 2.3 g/kg choline chloride instead of choline bitartrate; %Kcal: 19.3 protein, 64.0 carb, 16.7 fat). Dietary restriction (DR) was applied as a 30% food intake reduction as previously published23. On average, Ercc1∆/− mice are known to eat 2.3 g food per day. DR was initiated at 7 weeks of age with 10% food reduction (2.1 g/day), when animals reached almost maximum body weight and development was completed. Dietary restriction was increased weekly by 10%, until it reached 30% dietary restriction (1.6 g/day) from 9 weeks of age onward. DR was supplied around Zeitgeber time 10–12, 0–2 h before the active period, to limit alterations in the biological clock.
Protein restricted and macronutrient altered diets were provided AL starting at 4 (EMC) or 8 (RIVM) weeks of age, continuing for the remainder of life. All animals were randomized across diets/groups prior to the start of the interventions. Diets in the PR groups were all based on standard AIN93G synthetic pellets77 and were isocalorically compensated with cornstarch in a 1:1 ratio in RIVM cohorts. Additionally, the phosphorus content in the RIVM PR cohorts’ diets was compensated with KH2PO4. Protein restriction experiments in the RIVM (30–80%) were sweetened with an additional 50 g/kg dextrose. RIVM-based macronutrient-restricted diets were structured as defined in Fig. S2A. Diets were processed into pellets and irradiated with 9 kGy (Isotron, Ede, the Netherlands). Throughout all experiments mice had unrestricted access to water.
Phenotype scoring
The mice were weighed and visually inspected weekly and were scored in a blinded manner by at least two experienced research technicians for the onset of various phenotypical parameters. Whole-body tremors were scored if mice were trembling for a combined total of at least 10 s when put on a flat surface for 20 s. Impaired balance was determined by observing the mice walking on a flat surface for 20 s. Mice that had difficulties in maintaining an upright orientation during this period were scored as having imbalance.
RNA-sequencing and analysis
Snap-frozen liver tissues of 11-week-old Ercc1∆/− mice on diets with different macromolecular compositions (Supplementary Fig. 2A) were weighted to 30 mg and homogenized in QIAzol Lysis Reagent using a Qiagen tissuelyser LT. RNA was isolated using the Qiagen miRNeasy RNA isolation kit (QIAgen; 217004) following standard protocol with optional DNase (Qiagen, #79254) digestion step. RNA quality and quantity were assessed using the NanoDrop One (Thermo Fisher Scientific, ND-ONE-W) and Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA; G2939BA), respectively. Samples were sent for transcriptome sequencing (GenomeScan, Leiden, The Netherlands) on the Illumina NovaSeq 6000 platform. Analysis of raw data files was performed on our in house-generated data analysis pipeline. Removal of sequence adaptors from sequence reads was performed using Trimmomatic (version 0.39). Trimmed reads were aligned to mouse reference genome (annotation: gencode.vM20.annotation.gtf; genome: GRCm38.p6.genome.fa; http://gencodegenes.org/mouse/release_M20.html) using STAR (version 2.7.0 f). Read counts for each gene were obtained using FeatureCounts (as part of SubRead version 1.6.4), filtered using filterByExpr with count set to 10, and followed by TMM normalization and quantification of log fold changes and false discovery rates using EdgeR (version 3.32.1). All data files have been submitted to the NCBI gene expression omnibus (GEO accession number).
Ingenuity Pathway Analysis (IPA; Ingenuity Systems, QIAGEN) was used to analyse TMM normalized sequencing reads and find significantly altered upstream regulators and pathways. Upstream regulators were filtered for cytokines. For pathway analysis an absolute log2 fold-change cut-off of 0.5 was used to improve data interpretability. Functional tox analysis was run with standard settings and subsequently filtered for liver related processes.
For transcription stress analysis, all genes with a mean TMM value above 1 over all groups were kept. This resulted in ~13.500 genes. Genes were ordered by gene length and split into bins of 500. The percentage of upregulated genes in each bin and the mean gene length of genes per bin were reported.
GSEA (Gene set enrichment analysis) was performed in R (version 4.3.3) using the clusterProfiler package (version 4.10.0) on edgeR filtered data of all genes. Gene ranking was based on a gene significance score, or π-value, which was calculated for each gene by multiplying the log2(fold change) by -log10(p.adj) to retain significance and logFC directionality78. GSEA was ran using default settings, except for minGSSize and maxGSSize which were set to 1 and 5000 respectively. GSEA plots were made using enrichplot (version 1.22.0).
The Illumina NextSeq 500 full RNA sequence dataset on hypothalamus of 6-month-old C57BL/6 J wild-type mice which were kept for 12 weeks on diets with a different protein:carbohydrate ratio (diets D14071607 and D14071612 formulated by Research Diets, Inc.)51 was downloaded from NCBI GEO (GSE114960) and processed according to the above-described pipeline and transcription stress analysis with an additional 10k gene length cut-off to remove most non-coding transcripts ( ~ 16.500 genes of which ~11.500 > 10 kb). Additional microarray RNA expression datasets of livers from 15-month-old C57BL/6 J wild-type mice maintained on diets varying in protein, carbohydrate, and fat starting at 3 weeks of age (GSE85998)52, livers of 6-month-old wildtype Wistar male rats receiving a high protein or control diet for a duration of 4 months (GSE57858)53, and subcutaneous adipose tissue biopsies of overweight middle-aged human subjects adhered to a 25% energy restricted diet, either combined with high protein (1.5 g/kg-bw/d), or with standard protein (0.8 g/kg-bw/d) for 12 weeks (GSE84046)54, were all analysed with the online GEO2R-tool (Version info: R 4.2.2, Biobase 2.58.0, GEOquery 2.66.0, limma 3.54.0) using Benjamini & Hochberg (false discovery rate) adjustment to the P-values standard processing settings. Differentially expressed genes (pvalue ≤ 0.05 and logFC > |0.5 | ) were separated in up- and downregulated and plotted for their respective gene lengths.
Statistical analysis
All data with error bars are presented as mean ± standard deviation (SD), and the individual datapoints (dots) are presented in the bar graphs. All analyses were performed using IPA, GSEA, GraphPad Prism version 8.4.2 (GraphPad Software, La Jolla California USA) or excel. P-values expressed as *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001 were considered to be significant; when indicated as “ns”, comparisons should be considered non-significant.
Weekly body weights (per mouse) and food intake (per cage) from the age of intervention-start through the maximum age at which all groups of the respective experiment still had ≥55% survival, were included, excluding the weights of the animals that died during that period. Subsequently, all the body weights after the start of the intervention were log10-transformed, had the log10 values at the start of the intervention subtracted from them, and then, for convenience (this being no more than a normalization without any effect on the statistical tests), had the lowest log10-baseline weight of the entire dataset subtracted from them. The average food intake for week 5–8 was taken as a baseline when interventions started at 8 weeks of age. A baseline was set at 2.3 g when experiments started at 4 weeks of age. To this the average of all cages of a specific sex were added for convenience (this also had no effect on the statistical tests). Then, the area under the curve (AUC) was determined for body weight and food intake by the trapezoid rule for the entire selected time interval for each animal. Grubbs’ test in GraphPad Prism, with alpha = 0.01, was applied once to all AUC values of the experiment to identify and remove potential outliers. One-Way-ANOVA, with Dunnet’s multiple comparison test was used to estimate differences in experimental groups vs. control groups for bodyweight and food intake.
Statistics for lifespan and onset of phenotypes were performed by computing Kaplan–Meier estimates of the survival curves, estimating the log hazard ratio effect size with 95% confidence intervals, and comparing the survival functions of different groups with the log-rank test in GraphPad Prism.
Boxplot statistics for gene-length-dependent transcription decline were calculated using Mann-Whitney test based on differentially expressed genes. Significance in XY-scatterplots of gene length vs percentage up/down regulation were calculated using Spearman correlation analysis.
Data availability
All data files have been submitted to the NCBI gene expression omnibus (GEO accession number GSE293207).
References
Khavinson, V., Popovich, I. & Mikhailova, O. Towards realization of longer life. Acta Bio Med. Atenei Parm. 91, e2020054 (2020).
Scott, A. J., Ellison, M. & Sinclair, D. A. The economic value of targeting aging. Nat. Aging 1, 616–623 (2021).
Sander, M. et al. The challenges of human population ageing. Age Ageing 44, 185–187 (2015).
Kennedy, B. K. et al. Geroscience: linking aging to chronic disease. Cell 159, 709–713 (2014).
López-Otín, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. Hallmarks of aging: An expanding universe. Cell 186, 243–278 (2023).
Niedernhofer, L. J. et al. Nuclear genomic instability and aging. Annu Rev. Biochem. 87, 295–322 (2018).
Schumacher, B., Pothof, J., Vijg, J. & Hoeijmakers, J. H. J. The central role of DNA damage in the ageing process. Nature 592, 695–703 (2021).
Gyenis, A. et al. Genome-wide RNA polymerase stalling shapes the transcriptome during aging. Nat. Genet. 55, 268–279 (2023).
López-Otín, C. & Kroemer, G. Hallmarks of Health. Cell 184, 33–63 (2021).
Chatzidoukaki, O., Goulielmaki, E., Schumacher, B. & Garinis, G. A. DNA damage response and metabolic reprogramming in health and disease. Trends Genet. 36, 777–791 (2020).
López-Otín, C., Galluzzi, L., Freije, J. M. P., Madeo, F. & Kroemer, G. Metabolic control of longevity. Cell 166, 802–821 (2016).
Rajado, A. T. et al. How can we modulate aging through nutrition and physical exercise? An epigenetic approach. Aging 15, 3191–3217 (2023).
Oudmaijer, C. A. J., Komninos, D. S. J., Hoeijmakers, J. H. J., IJzermans, J. N. M. & Vermeij, W. P. Clinical implications of nutritional interventions reducing calories, a systematic scoping review. Clin. Nutr. ESPEN 63, 427–439 (2024).
Green, C. L., Lamming, D. W. & Fontana, L. Molecular mechanisms of dietary restriction promoting health and longevity. Nat. Rev. Mol. Cell Biol. 23, 56–73 (2022).
Morales-Suarez-Varela, M., Collado Sánchez, E., Peraita-Costa, I., Llopis-Morales, A. & Soriano, J. M. Intermittent fasting and the possible benefits in obesity, diabetes, and multiple sclerosis: a systematic review of randomized clinical trials. Nutrients 13, 3179 (2021).
Mazandarani, M., Lashkarbolouk, N., Ejtahed, H. S. & Qorbani, M. Does the ketogenic diet improve neurological disorders by influencing gut microbiota? A systematic review. Nutr. J. 22, 61 (2023).
De Cabo, R. & Mattson, M. P. Effects of intermittent fasting on health, aging, and disease. N. Engl. J. Med. 381, 2541–2551 (2019).
Lee, M. B., Hill, C. M., Bitto, A. & Kaeberlein, M. Antiaging diets: Separating fact from fiction. Science 374, eabe7365 (2021).
Speakman, J. R. & Mitchell, S. E. Caloric restriction. Mol. Asp. Med. 32, 159–221 (2011).
Fontana, L., Partridge, L. & Longo, V. D. Extending Healthy Life Span—From Yeast to Humans. Science 328, 321–326 (2010).
Kraus, W. E. et al. 2 years of calorie restriction and cardiometabolic risk (CALERIE): exploratory outcomes of a multicentre, phase 2, randomised controlled trial. Lancet Diab. Endocrinol. 7, 673–683 (2019).
Dorling, J. L. et al. Effects of caloric restriction on human physiological, psychological, and behavioral outcomes: highlights from CALERIE phase 2. Nutr. Rev. 79, 98–113 (2021).
Vermeij, W. P., et al. Restricted diet delays accelerated ageing and genomic stress in DNA-repair-deficient mice. Nature 537, 427–431 (2016).
Birkisdóttir, M. B. et al. The use of progeroid DNA repair-deficient mice for assessing anti-aging compounds, illustrating the benefits of nicotinamide riboside. Front Aging 3, 1005322 (2022).
Gurkar, A. U. & Niedernhofer, L. J. Comparison of mice with accelerated aging caused by distinct mechanisms. Exp. Gerontol. 68, 43–50 (2015).
Marteijn, J. A., Lans, H., Vermeulen, W. & Hoeijmakers, J. H. J. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell Biol. 15, 465–481 (2014).
Dollé, M. E. T. et al. Broad segmental progeroid changes in short-lived Ercc1 −/Δ7 mice. Pathobiol. Aging Age-Relat. Dis. 1, 7219 (2011).
Vermeij, W. P., Hoeijmakers, J. H. J. & Pothof, J. Genome integrity in aging: human syndromes, mouse models, and therapeutic options. Annu Rev. Pharm. Toxicol. 56, 427–445 (2016).
Borgesius, N. Z. et al. Accelerated age-related cognitive decline and neurodegeneration, caused by deficient DNA repair. J. Neurosci. 31, 12543–12553 (2011).
De Waard, M. C. et al. Age-related motor neuron degeneration in DNA repair-deficient Ercc1 mice. Acta Neuropathol. ((Berl.)). 120, 461–475 (2010).
Arvanitaki, E. S. et al. Microglia-derived extracellular vesicles trigger age-related neurodegeneration upon DNA damage. Proc. Natl. Acad. Sci. 121, e2317402121 (2024).
Bruce, K. D. et al. High carbohydrate–low protein consumption maximizes Drosophila lifespan. Exp. Gerontol. 48, 1129–1135 (2013).
Tatar, M., Post, S. & Yu, K. Nutrient control of Drosophila longevity. Trends Endocrinol. Metab. 25, 509–517 (2014).
Solon-Biet et al. The ratio of macronutrients, not caloric intake, dictates cardiometabolic health, aging, and longevity in Ad Libitum-Fed Mice. Cell Metab. 19, 418–430 (2014).
Wahl, D. et al. Comparing the effects of low-protein and high-carbohydrate diets and caloric restriction on brain aging in mice. Cell Rep. 25, 2234–2243.e6 (2018).
Birkisdóttir, M. B. et al. Unlike dietary restriction, rapamycin fails to extend lifespan and reduce transcription stress in progeroid DNA repair-deficient mice. Aging Cell. 20, e13302 (2021).
Leto, S., Kokkonen, G. C. & Barrows, C. H. Dietary protein, life-span, and physiological variables in female mice. J. Gerontol. 31, 149–154 (1976).
Lagiou, P. et al. Low carbohydrate-high protein diet and incidence of cardiovascular diseases in Swedish women: prospective cohort study. BMJ 344, e4026–e4026 (2012).
Zhang, X. et al. Identification of a leucine-mediated threshold effect governing macrophage mTOR signalling and cardiovascular risk. Nat. Metab. 6, 359–377 (2024).
Lennerz, B. S., Koutnik, A. P., Azova, S., Wolfsdorf, J. I. & Ludwig, D. S. Carbohydrate restriction for diabetes: rediscovering centuries-old wisdom. J. Clin. Invest. 131, e142246 (2021).
Sun, J. et al. The effect of dietary carbohydrate and calorie restriction on weight and metabolic health in overweight/obese individuals: a multi-center randomized controlled trial. BMC Med. 21, 192 (2023).
Milanese, C., et al. DNA damage and transcription stress cause ATP-mediated redesign of metabolism and potentiation of anti-oxidant buffering. Nat. Commun. 10, 4887 (2019).
Liu, Y., Munker, S., Müllenbach, R. & Weng, H. L. IL-13 signaling in liver fibrogenesis. Front Immunol. 3, 116 (2012).
Yuan, Y. et al. Leukemia inhibitory factor protects against liver steatosis in nonalcoholic fatty liver disease patients and obese mice. J. Biol. Chem. 298, 101946 (2022).
Weng, S. Y. et al. IL-4 Receptor alpha signaling through macrophages differentially regulates liver fibrosis progression and reversal. EBioMed. 29, 92–103 (2018).
Dinarello, C. A. Overview of the IL -1 family in innate inflammation and acquired immunity. Immunol. Rev. 281, 8–27 (2018).
Gulen, M. F. et al. cGAS–STING drives ageing-related inflammation and neurodegeneration. Nature 620, 374–380 (2023).
Stoeger, T. et al. Aging is associated with a systemic length-associated transcriptome imbalance. Nat. Aging 2, 1191–1206 (2022).
Barbash, S. & Sakmar, T. P. Length-dependent gene misexpression is associated with Alzheimer’s disease progression. Sci. Rep. 7, 190 (2017).
Soheili-Nezhad, S., Ibáñez-Solé, O., Izeta, A., Hoeijmakers, J. H. J. & Stoeger, T. Time is ticking faster for long genes in aging. Trends Genet 40, 299–312 (2024).
Hu, S. et al. Dietary fat, but not protein or carbohydrate, regulates energy intake and causes adiposity in mice. Cell Metab. 28, 415–431.e4 (2018).
Gokarn, R., et al. Long-term dietary macronutrients and hepatic gene expression in aging mice. J. Gerontol. A Biol. Sci. Med. Sci. 73, 1618–1625 (2018).
Díaz-Rúa R, Keijer, J., Palou, A., van Schothorst, E. M. & Oliver, P. Long-term intake of a high-protein diet increases liver triacylglycerol deposition pathways and hepatic signs of injury in rats. J. Nutr. Biochem. 46, 39–48 (2017).
Van Bussel, I. P. G. et al. The impact of protein quantity during energy restriction on genome-wide gene expression in adipose tissue of obese humans. Int J. Obes. 2005. 41, 1114–1120 (2017).
Solon-Biet, S. M. et al. Macronutrient balance, reproductive function, and lifespan in aging mice. Proc. Natl. Acad. Sci. 112, 3481–3486 (2015).
Kitada, M., Ogura, Y., Monno, I. & Koya, D. The impact of dietary protein intake on longevity and metabolic health. EBioMedicine 43, 632–640 (2019).
Green, C. L. et al. Sex and genetic background define the metabolic, physiologic, and molecular response to protein restriction. Cell Metab. 34, 209–226.e5 (2022).
Pak, H. H. et al. Fasting drives the metabolic, molecular and geroprotective effects of a calorie-restricted diet in mice. Nat. Metab. 3, 1327–1341 (2021).
Mirzaei, H., Suarez, J. A. & Longo, V. D. Protein and amino acid restriction, aging and disease: from yeast to humans. Trends Endocrinol. Metab. Tem. 25, 558–566 (2014).
Weigle, D. S. et al. A high-protein diet induces sustained reductions in appetite, ad libitum caloric intake, and body weight despite compensatory changes in diurnal plasma leptin and ghrelin concentrations. Am. J. Clin. Nutr. 82, 41–48 (2005).
Lopes, I., Altab, G., Raina, P. & De Magalhães, J. P. Gene size matters: an analysis of gene length in the human genome. Front Genet. 12, 559998 (2021).
Le Couteur, D. G. et al. The impact of low-protein high-carbohydrate diets on aging and lifespan. Cell Mol. Life Sci. 73, 1237–1252 (2016).
Harman, D. Aging: a theory based on free radical and radiation chemistry. J. Gerontol. 11, 298–300 (1956).
Tang, R. et al. Recent progress in the effect of ferroptosis of HSCs on the development of liver fibrosis. Front Mol. Biosci. 10, 1258870 (2023).
Du, K. et al. Aging promotes metabolic dysfunction-associated steatotic liver disease by inducing ferroptotic stress. Nat Aging 4, 949–968 (2024).
Mulderrig, L. et al. Aldehyde-driven transcriptional stress triggers an anorexic DNA damage response. Nature 600, 158–163 (2021).
Wang, M. et al. Genotoxic aldehyde stress prematurely ages hematopoietic stem cells in a p53-driven manner. Mol. Cell. 83, 2417–2433.e7 (2023).
Ukraintseva, S. et al. Decline in biological resilience as key manifestation of aging: Potential mechanisms and role in health and longevity. Mech. Ageing Dev. 194, 111418 (2021).
Akter, S. et al. Low carbohydrate diet and all cause and cause-specific mortality. Clin. Nutr. Edinb. Scotl. 40, 2016–2024 (2021).
Noto, H., Goto, A., Tsujimoto, T. & Noda, M. Low-carbohydrate diets and all-cause mortality: a systematic review and meta-analysis of observational studies. PloS One 8, e55030 (2013).
Floegel, A. & Pischon, T. Low carbohydrate-high protein diets. BMJ 344, e3801–e3801 (2012).
Levine, M. E. et al. Low protein intake is associated with a major reduction in IGF-1, cancer, and overall mortality in the 65 and younger but not older population. Cell Metab. 19, 407–417 (2014).
Landry, M. J. et al. Cardiometabolic effects of omnivorous vs vegan diets in identical twins: a randomized clinical trial. JAMA Netw. Open. 6, e2344457 (2023).
Song, M. et al. Association of animal and plant protein intake with all-cause and cause-specific mortality. JAMA Intern Med. 176, 1453 (2016).
Ardisson Korat, A. V. et al. Dietary protein intake in midlife in relation to healthy aging – results from the prospective Nurses’ Health Study cohort. Am. J. Clin. Nutr. 119, 271–282 (2024).
Weeda, G. et al. Disruption of mouse ERCC1 results in a novel repair syndrome with growth failure, nuclear abnormalities and senescence. Curr. Biol. 7, 427–439 (1997).
Reeves, P. G. Components of the AIN-93 Diets as Improvements in the AIN-76A Diet. J. Nutr. 127, 838S–841S (1997).
Xiao, Y. et al. A novel significance score for gene selection and ranking. Bioinformatics 30, 801–807 (2014).
Acknowledgements
We are grateful to Bhawani Nagarajah and the animal caretakers for general assistance with mouse experiments. This research was funded by the National Institute of Health (NIH)/National Institute of Ageing (NIA) (AG17242), the National Institute for Public Health and the Environment and the Ministry of Health, Welfare and Sport of The Netherlands (S/132002), and ONCODE supported by the Dutch Cancer Society. J.H.J.H. was additionally supported by European Research Council Advanced Grant Dam2Age, and Deutsche Forschungsgemeinschaft (DFG, German Research Foundation—Project-ID 73111208—SFB 829); J.H.J.H. and W.P.V. by Memorabel (ZonMW 733050810), BBoL (NWO-ENW 737.016.015), and the European Joint Programme Rare Diseases (TC-NER RD20-113); and W.P.V. by the Regiodeal Foodvalley (162135) and ADPS Longevity Research Award.
Author information
Authors and Affiliations
Contributions
IvG, JHJH, METD, and WPV conceptualized and designed the experiments, performed statistical analysis of data, and wrote the manuscript. MBB, RMCB, SB, SI, CTvO, KS, YMAR, ER, HvS, METD, and WPV planned and performed mouse experiments and behavioural assessments. IvG, RAO, RWGNvdM, and WPV performed transcriptome analysis. All authors contributed to the manuscript and read and approved the final submitted version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
van Galen, I., Birkisdóttir, M.B., Ozinga, R.A. et al. High protein intake causes gene-length-dependent transcriptional decline, shortens lifespan and accelerates ageing in progeroid DNA repair-deficient mice. npj Metab Health Dis 3, 20 (2025). https://doi.org/10.1038/s44324-025-00064-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s44324-025-00064-3
This article is cited by
-
Aging-Related Obesity: Unveiling Mitochondrial and Metabolic Dysfunction
Current Nutrition Reports (2025)