Abstract
A novel method uses buoyant microbubbles and magnetic microspheres to label, separate, and detect SARS-CoV-2 N-protein in patient saliva at the best reported limit of detection to date. The equipment needed is remarkably simple and inexpensive: a flashlight, digital camera, magnet, and cuvette holder, which facilitates point-of-care deployment. In our method, saliva is mixed with lysis buffer, antibody-functionalized ~15 µm buoyant gas-filled silica microbubbles, and 2.7 µm antibody-functionalized polystyrene magnetic microspheres, forming buoyant-analyte-magnetic (BAM) complexes. A magnet pulls the BAM complexes to the bottom of the cuvette while unbound microbubbles float upwards. Removing the magnet releases the buoyant BAM complexes, which appear as bright rising dots under flashlight illumination. A camera counts the BAM complexes, with an analytical detection limit of ~37 SARS-CoV-2 N-protein molecules in 5 µL of saliva. The assay provided positive results for all tested PCR-positive saliva specimens, with concentrations ranging from 0.7 to 2.5 × 105 RNA copies/µL.

Similar content being viewed by others
Detecting infectious diseases such as COVID-19 early and at the point of care can improve treatment outcomes and prevent transmission. This is especially true during outbreaks and pandemics, which can have severe mortality1, morbidity, social, and economic repercussions. However, at early stages, patient specimens contain only minute concentrations of pathogen-specific biomarkers, which makes early detection challenging2,3,4,5. Additionally, point-of-care assays should ideally adhere to the World Health Organization’s (WHO)’s ASSURED criteria for resource-constrained diagnostics (Affordable, Sensitive, Specific, User-friendly, Rapid/Robust, Equipment-free or battery-powered, and Deployable)6,7. While each application will have its own quantitative and qualitative targets for these criteria, there is often a choice between ultrasensitive assays (which usually require multiple steps and sensitive equipment) and point-of-care detection.
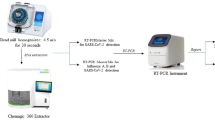
Unfortunately, most assays for Covid-19 and many other diseases are either insufficiently sensitive or unsuitable for point-of-care detection (Fig. 1). The WHO’s 2020 Target Product Profile (TPP) for Covid-19 diagnostics8 gives target limits of detection and assay time for both point-of-care applications (acceptable target of <103 RNA cp/µL in 40 min; desirable target of <10 cp/µL in 20 min) and for diagnostic/confirmation applications (1 cp/µL in 4 hr acceptable; 0.1 cp/µL in 45 min desirable). Generally, qPCR meets the ASSURED sensitive and specific criteria and WHO TPP criteria for diagnostics, but is insufficiently affordable, user-friendly, rapid, equipment-free, or deployable for many point-of-care applications9. By contrast, rapid lateral flow assays are available in pharmacies10,11 and are suitable for home and point-of-care use, but their sensitivity is relatively low, with frequent real-world false negatives (~50% sensitivity for asymptomatic patients and 70% for symptomatic COVID-19 patients)12. Most commercialized lateral flow assays have detection limits that fall within the range equivalent to 104 to 102 cp/µL13,14,15, with many tests meeting the adequate WHO point-of-care TPP specification, but not the desirable specification, and all are insufficient for diagnostic needs. Consequently, these tests often miss infections at potentially infectious levels (especially with prolonged close contact)16,17. More sensitive lateral flow assays can be achieved using specialized nanoparticle labels, including magnetic capture18 and single-molecule scanning with SERS19, but these require specialized instrumentation to read them. Portable COVID-19 isothermal nucleic acid amplification tests have been commercialized, such as Abbott’s ID Now test. Unfortunately, these devices and assays are moderately expensive and have a reported detection limit of 3 × 102 RNA copies/µL, which can still be infectious17. Thus, more sensitive and faster assays are needed when definitive diagnosis is required (e.g., urgent care facilities)20, to prevent transmission to vulnerable and immunosuppressed patients (e.g., in skilled nursing facilities)21, and to more effectively contain disease outbreaks22,23.
The most sensitive antigen tests available label and count single molecules24. The analyte is often sandwiched between two types of antibody-functionalized labels: magnetic microspheres and a detection label. The reaction is reasonably rapid because most analyte diffuses only short distances to reach the microspheres, unlike diffusion to flat surfaces in traditional microwell immunoassays25. The magnetic microspheres are then magnetically collected and rinsed to remove unbound detection label. Any detection label still bound to the magnetic microspheres is then enzymatically developed26,27. For example, David Walt’s group developed a single molecule array (Simoa) assay, commercialized by Quanterix, which captures analyte using magnetic microspheres and labels this analyte with fluorogenic enzymes28,29. Similarly, Chad Mirkin’s group developed bio-barcode assays using magnetic microspheres and DNA-functionalized gold nanoparticles as the detection label, with PCR/gene chip or silver development as a readout30. However, the required rinsing and development steps in these protocols increase time, protocol complexity, instrument complexity, and cost, which renders them unsuitable for rapid, point-of-care detection.
Like the Simoa and biobarcode assays, our buoyant-analyte-magnetic (BAM) assay captures analyte molecules with both magnetic particles and a detection label; however, our detection labels are ~15 µm gas-filled silica-shell buoyant microbubbles (Fig. 2), which separate from unbound labels without rinsing and are easily detected without development steps. The microbubbles have four key advantages: First, they rapidly sample a large solution volume as they rise in solution (~2 mm/min), providing rapid and efficient analyte and magnetic microsphere capture31. Similarly, buoyant microbubbles have been used for high capture efficiency, simple cell separation32, including the separation and detection of Escherichia coli33, and ultrasensitive electrochemical depletion assays34. Second, the unbound microbubbles rise away from the bottom surface during magnetic separation, which clears the background solution in minutes without rinsing. Third, microbubbles and BAM complexes scatter light from a flashlight so intensely that they can be observed by the naked eye and counted using inexpensive portable cameras. Similarly, Chorti and co-workers described an immunoassay where analyte molecules bound to both 100 nm diameter magnetic nanospheres and a single slightly buoyant 1 mm diameter polystyrene sphere in a salt solution: when enough analyte molecules and magnetic nanospheres bound (typically > 106), the polystyrene sphere could be pulled down with a magnet as observed by eye35. We apply the principle more sensitively: one magnetic microsphere provides enough force to pull down a ~15 µm buoyant microbubble, and we count how many of the 750,000 microbubbles in our reaction mixture bind to magnetic microspheres. Finally, opposing buoyant and magnetic forces reduce nonspecific binding by pulling apart weakly bound BAM complexes and keeping them separated. Together, these features enable rapid single-molecule-counting using inexpensive portable equipment.
Results
We first calibrated the BAM assay using simulated saliva samples. We then performed qualitative and quantitative analysis in patient saliva specimens and compared results to saliva RT-qPCR.
Simulated saliva studies
Simulated saliva was spiked with varying N-protein concentrations, and the BAM complexes were counted. The nonspecific background with no N-protein was 12.5 ± 8.8 BAM complexes. Figure 3 shows a good linear regression fit (R2 = 0.99). The slope, m = 0.84 BAM complexes/N-protein, implies an 84% analyte capture-and-detection efficiency. The analytical limit of detection (LOD) was 31 N-proteins, given by 3sy/m, where sy is the noise on the blank (8.8 BAM complexes). This is equivalent to 0.24 fg/mL in 10 μL 10% simulated saliva, or 2.4 fg/mL in the original 1 µL saliva before 10× dilution.
An average of 0, 64, 127, 255, 764 and 1528 N-protein molecules were spiked into simulated saliva and resulting BAM complexes were counted. At >255 molecules/µL, BAM complexes were hard to count directly due to overlapping tracks; instead, BAM complexes were counted in \(1/10\) of the mixture and multiplied by 10×. Each point shows the average and standard deviation of 4 trials.
We also made several qualitative observations: Within 15 min of applying the magnetic field, all magnetic particles were at the bottom of the cuvette, and most of the buoyant microbubbles had risen off the bottom, except for some occasional low-buoyancy microbubbles and debris. We also determined that surface chemistry is critical to maintaining low background levels, as much more background was observed without biotin-PEG functionalization (Supplementary Fig. 1). Additionally, although not part of the standard protocol, we found that most of the BAM complexes were intact and could be pulled back to the cuvette bottom by reapplying the magnet. This happened even for non-specifically bound complexes (with no added N-protein), however, this background decreased with each pulldown as some particles spontaneously pulled apart (Supplementary Fig. 2). We observed that when the magnet was removed and then immediately replaced, a small fraction of BAM complexes were not recollected. These tracks are likely from BAM complexes that break apart when we remove the magnet, which can induce forces and torques on chained magnetic particles; we also cannot rule out that they are from free buoyant beads that are temporarily buried beneath magnetic particle chains, though thermodynamically we would expect magnetic particles would push non-magnetic particles out rather than trapping them. Regardless, the fluctuations in the quantity of this incidental phenomenon will be reflected in the background signals, allowing us to subtract this signal during the data processing.
In addition, we observed variable track thicknesses, attributable to the buoyant microbubble size range (5–35 µm). Furthermore, large aggregates with buoyant microbubbles were occasionally noted in negative tests with non-specific binding. Aggregates were also observed in both the high concentration spike tests and high concentration patient saliva assays.
Lastly, the detected number of BAM complexes increased with incubation time and fit well to an exponential regression with an 8-min time constant (Supplementary Fig. 3 and Table 1). Our standard protocol used a 30-min incubation to ensure high capture efficiency and negligible dependence on reaction time variation.
Spiked PCR-negative patient saliva assays
In PCR-negative patient saliva, the BAM assay gave a nonspecific background of 77 ± 10 BAM complexes (Supplementary Fig. 4). This background, although relatively low, was significantly larger than the background in simulated saliva, which is likely due to mucin and other molecules found in saliva. Diluting the saliva by 2×, 10×, and 100×, respectively, reduced the background to 36 ± 3, 29 ± 2, and 19 ± 4 BAM counts (Supplementary Fig. 4). Capture-and-detection efficiency was 78% in saliva samples diluted to 1% concentration (Supplementary Fig. 5D) and 91% in samples diluted to 50% concentration (Supplementary Fig. 6D). Since 2× dilution (i.e., 5 µL patient saliva diluted to 10 µL) significantly reduced the PCR-negative background while maintaining high capture efficiency, all subsequent experiments employed 2× dilution.
Positive and negative patient studies
Next, we performed BAM assays on n = 3 PCR-positive and n = 8 PCR-negative patient saliva specimens. As shown in Fig. 4A, the positive specimen is clearly distinguishable from the negative specimen by the significant difference in the number of BAM tracks. Across all tested specimens (Fig. 4B), the average number of BAM complexes in positive samples was 411 ± 151, compared to 19 ± 10 in negative samples. The positive and negative specimens were completely distinguished (p = 5 × 10−5) with no negative specimens reading over 49 BAM counts and no positive specimens reading below 208. Supplementary Fig. 7 and Table 2 show the same data plotted as sensitivity and specificity versus threshold, as well as a ROC curve.
Serial dilution and quantification
The addition of concentrated N-protein solution to positive patient saliva caused no evident increase in BAM counts, and diluting the patient saliva specimen initially caused the BAM counts to increase before eventually decreasing with further dilution (see Supplementary Fig. 8). This is consistent with the hook effect, which states that very high levels of analyte can lead to a paradoxical decrease in assay signal. This result illustrates that the assay loses linearity at high N-protein concentrations.
To address the difficulties of counting >~250 BAM complexes due to overlapping tracks, and non-linearities from aggregation, and surface saturation, we diluted each specimen to the linear range for quantification. Further diluting the diluted PCR-positive samples by an additional 100× (up to 108-fold overall), resulted in 21 ± 7 BAM complexes (Supplementary Table 3), essentially matching the negative background from PCR-negative samples (19 ± 10 BAM complexes, Fig. 4B).
Performance at different temperatures
To determine whether the test needed a temperature-controlled environment or could be performed in a range of outdoor temperatures, we performed spike-recovery BAM assay in 10% simulated saliva and 50% patient saliva as well as low-positive specimens at 5 °C, room temperature (18–20 °C), and 40 °C (see Supplementary Fig. 9 and Table 4). Neither temperature nor sample type significantly affected detection efficiency, and pooled data had a range of 99–127 additional BAM complexes over background per ~127 molecules added (on average), implying 78–100% capture efficiency, essentially consistent with our previous measurements of 78–91%. We did see a significantly higher background at 40 °C (29 ± 14 in simulated saliva and 54 ± 13 in real saliva) than either room temperature (9 ± 6 in simulated saliva; 18 ± 2 in real saliva) or 5 °C (9 ± 6 in simulated saliva; 21 ± 15 in real saliva). The negative real saliva average value (54 counts) and the signal-LOD calculated from the four negative trials at 40 °C was 93 BAM counts. These were above the prior signal-LOD of the room temperature test (50 counts), although still well below the lowest positive value observed in Fig. 4B data (208). Importantly, positive patient saliva from patients with low viral loads after symptoms resolved (<1 cp/µL by PCR) were clearly positive for all trials under all three temperature conditions (n = 4 trials per condition). Together, these results show that the test can be performed in a range of conditions with similar capture efficiency, although the positive-threshold should be set above the room temperature LOD.
After accounting for the dilution factor, the BAM concentrations were compared to RNA concentrations calculated from Ct values (Fig. 5). The linear regression slope is ~470 BAM complexes per RNA copy (Fig. 5A). The linear relationship between BAM complexes and RNA is maintained over a wide concentration range, from <1 cp/µL to 25000 cp/µL, levels high enough to be detectable by lateral flow assays (Fig. 5B).
PCR-measured RNA copies/μl for n = 8 PCR positive and n = 8 PCR negative patient saliva specimens. Although all positive specimens gave positive BAM results at initial concentration, overlapping tracks prevented quantification; thus, for quantification, specimens were diluted by up to 106× to the linear assay-response region. A Plot on linear x- and y-axis scales. B Plot from A on a log y-scale and PCR Ct value x-scale. The linear regression slope was ~470 BAM complexes/RNA. The signal LOD (3sy + b) from negative specimens was 50 BAM complexes in 5 μL patient saliva.
Discussion
The BAM assay was designed for the ultrasensitive, rapid, and affordable detection of early-stage diseases. Below, we discuss the assay’s LOD, quantification, protocol time, affordability, and deployability/user-friendliness.
The analytical signal LOD for the assay using 5 µL patient saliva can be estimated as 3sy/m, where sy is the noise on the blank (10.4 BAM complexes from Fig. 4B) and m is the calibration curve slope, (0.91 BAM/N-protein in saliva, Supplementary Fig. 6). The analytical signal LOD for the patient saliva assay at room temperature was 37 N-protein molecules per 5 µL, or 0.29 fg/mL, or 12 aM (with about 18% uncertainty estimated from variation in sy and m in different experiments). To the best of our knowledge, this is the lowest LOD for N-protein concentration in patient saliva reported to date. For example, a commercial N-protein Quanterix Simoa assay has a reported LOD of 99 fg/mL in nasopharyngeal swabs diluted in 100 µL buffer36, and an in-house Simoa assay was reported with an LOD of 20 fg/mL37. Other sandwich immunoassays with magnetic separation for other proteins have LODs almost as good. For example, Nam et al. report a 30 aM LOD for human prostate-specific antigen in 10 µL of goat serum using a biobarcode sandwich assay with a DNA-detection label and PCR as a readout30. However, the BAM assay avoids rinsing and development steps, which simplifies protocols, reduces total assay time, and reduces instrumentation cost, complexity, and size.
The BAM assay’s ultralow LOD is due to both high capture-and-detection efficiency (in 30 min, 78–91% capture-and-detection efficiency according to Fig. 3, Supplementary Figs. 3, 5D, and 6D) and low noise from variation in blank samples. Each BAM complex scatters light intensely, enabling high detection efficiency. Avoiding rinsing, nanowell loading, and development steps also reduces analyte loss. The high analyte capture efficiency could be explained by the large volume explored by the microbubbles as they rise. For example, literature on planktonic feeding shows that rising at ~50 µm/s increases the analyte molecule capture rate by ~2× compared to diffusion38, and greatly increases the rate of subsequent capture of magnetic microspheres (which have a large impact cross-section but diffuse slowly due to their size)39,40.
While we have not directly imaged and counted analyte molecules in a BAM complex, two arguments imply that at low concentrations, each BAM complex contains one analyte molecule (aside from non-specifically bound complexes). First, we theoretically expect that the number of N-protein molecules/buoyant microbubble should follow a Poisson distribution: P(k) = (λk/e−λ)/k!, where P(k) is the probability that exactly k molecules bind to a given bead and λ is the average number of captured molecules/bead. At low concentrations, almost all beads have 0 or 1 molecules even if every molecule is captured (e.g., for 1 fg/mL, λ = 127/750,000 molecules/microbubbles and only one in 70,000,000 microbubbles captures two or more analyte molecules; even for λ = 50%, only 9% of microbubbles capture two or more molecules). However, at higher concentrations, aggregates of multiple molecules/bead are expected and observed (e.g., Supplementary Fig. 8). Second, a BAM complex can represent at most one analyte molecule (because N-proteins are 1000x smaller than the magnetic particles and do not self-replicate); there can be <1 BAM complex/molecule if n molecules bind to one buoyant microbubble (in which case the capture efficiency could at most be 1/n), or if not all molecules are captured and detected. Our observed high capture and detection efficiency at low concentrations (78–91%) indicates that BAM complexes at these concentrations largely correspond to single molecules: Assuming an 85% efficiency, if all the efficiency loss came from BAM complexes with 2 N-proteins, 82% of BAM complexes would be from single molecules and 18% from double. However, combining the arguments, the Poisson statistics make even the 18% double-analyte BAM complexes unlikely at low concentration, implying most of the inefficiency comes from imperfect capture or detection. Regardless, our assay has very high sensitivity and efficiency.
In addition to efficiently capturing and detecting most analyte molecules, the excellent LOD arises from low nonspecific binding background variation. The background from n = 8 negative patients (24 tests) was 19 ± 10 (Fig. 5B), and the background from the same number of positive specimens, after diluting to close to 0 concentration (up to 108-fold dilution) was 21 ± 7 (Supplementary Table 3). The total background counts are low, and variation is correspondingly low, although it was higher than shot noise and included variation from multiple specimens, days, and lots. Moreover, the assay consistently detects small numbers of molecules, including in calibration curves (Supplementary Figs. 5D and 6D), the reaction rate curve (Supplementary Fig. 3 and Table 1), and correctly identified 8 negative specimens and 3 positive patient specimens in Fig. 4B (in each of the triplicate trials, 33 trials overall), another n = 8 positive specimens during initial undiluted assessment in serial dilution study Fig. 5 (and continuing positive in samples diluted by up to a factor of 106), and n = 12 trails (4 trials/temperature) from one PCR positive patient in the temperature studies with n = 24 trials for the negative and spiked-negative specimens (Supplementary Fig. 9). More studies are planned after we develop the next-generation portable prototypes and rapid protocols.
Mechanistically, two factors contribute to the low nonspecific binding background. First, good surface chemistry is essential (e.g., blocking free streptavidin sites with biotin-PEG, see Supplementary Fig. 1A). Second, the opposing buoyant and magnetic forces can break apart weak nonspecific interactions. A 15 µm microbubble with a 0.6 g/cm2 density has a buoyancy force in buffer of ~7 pN. Since the magnet pulls down BAM complexes ~5× as quickly as they rise, we expect that during this process the magnetic and buoyant forces are pulling on the N-protein with ~6× as much force (~40 pN), albeit for the brief period (~30 s) when the BAM complexes approach the bottom of the cuvette but before reached it. Previous studies on pulling apart bonds with AFM tips, magnetic tweezers, centrifugation, and optical tweezers found that the dissociation time decreases exponentially with force, roughly in accordance with Bell’s model24,26,27. The forces and times calculated here are sufficient to keep apart spontaneously-dissociating particles and break apart weak interactions, but over the short experiment, are unlikely to affect antibody-antigen interactions with high KD (e.g., 10 pM for the R040 anti-N-protein antibody) and force-free dissociation times of days to months41,42. The forces were sufficient to reduce nonspecific binding while retaining high capture efficiency; in the future, the magnetic forces, bead sizes, and surface chemistry configurations can be further optimized.
N-protein concentrations were quantified by diluting the saliva specimen to the linear region (and accounting for background and dilution factor). This approach is analogous to how bacteria are traditionally quantified by diluting until individual bacterial colony-forming units can be counted (or similarly, plaque-forming units for viruses). The slope of Fig. 4B was 470 BAM complexes per RNA copy, or around 500 N-proteins/RNA, assuming an 84% capture efficiency. This agrees with electron microscopy of individual (pleomorphic) virions giving 38 ± 10 ribonucleic acid complexes per virion, with simulations showing 12 N-proteins per ribonucleic acid complex, giving a total of 456 ± 120 N-proteins/virion43,44. However, we are measuring N-protein in the whole saliva specimen, not necessarily from within intact virions, and in different conditions (after Triton-X addition for N-protein detection, and after baking for RNA RT-qPCR detection). The measured concentrations could be affected by protease- and nuclease-degradation of N-proteins and RNA, respectively, antibodies masking the N-proteins, and measurement uncertainties in both BAM and RT-qPCR assays. To further illustrate this point, other studies found >10x specimen-to-specimen N-protein variation at the same Ct value36,37. Regardless, the dilution approach provided a robust quantitative analysis over a wide range of concentrations (up to 108 BAM/5 µL saliva).
Admittedly, the serial dilution steps used for quantification reduce assay speed, user-friendliness, and affordability. If needed, we expect that dilution can be automated with microfluid systems, with different regions in one or more cuvettes used to assess different concentrations45. Additionally, for faster assays (when capture efficiency becomes strongly time-dependent), ratiometric measurements could be performed by multiplexing based on microbubble size, fluorescence, or chemically or photocleavage release time46. Diluting the specimen and counting also inevitably keeps the total number of counted BAM complexes low, where Poisson statistics (shot noise) limits relative noise to 1/sqrt(n) (e.g., 10% for 100 BAM complexes). If needed, in the future, a wider dynamic range might be achieved by spreading the beads out over a larger area instead of having them rise from a single line, or by more complex analysis to look at bead aggregation or intensity. However, for most infectious diseases, quantification is unnecessary and qualitative infection status results are sufficient, especially for rapid on-site tests. The current assay protocol was designed to show proof-of-principle, and we used a relatively long incubation time (30 min) to capture most analytes in the specimen, which improved reproducibility and illustrated an excellent LOD. However, 30 min would be too long for many point-of-care applications. Fortunately, we found that the BAM formation time constant was 8 min (Supplementary Fig. 3 and Table 1). Thus, theoretically, a 1-min incubation time should provide 10% capture efficiency, allowing manual mixing (e.g., by squeezing a tube), which would be sufficient for many applications.
The total assay time also includes time for saliva collection, magnetic separation, and readout with the magnet removed. The collection step was ~4 min, including Salivette centrifugation, but could be sped up, especially for small 5 µL specimens. A 15-min magnetic separation time provided a clear read-out zone at the bottom of the cuvette. However, in future, removing the small/slowly-rising microbubbles would accelerate this time. Finally, up to 10 min of video was recorded after magnet removal to analyze BAM particle motion. However, >50% of the BAM complexes were read within the first 10 frames (20 s), (see Supplementary Fig. 10 and Table 5). Thus, although the total assay time was around 55 min, we expect that it could be reduced to ~3–15 min, with some loss in LOD, but still unprecedented sensitivity for a rapid portable immunoassay.
To meet the WHO’s ASSURED criteria, the equipment and reagents must all be affordable. The most expensive and heaviest component is the Nikon D7000 DLS camera. However, since the particles are naked-eye visible, most digital cameras will work, and indeed, we found that a phone camera with no additional optics (Samsung Galaxy Z Flip6) could easily detect 1 fg/mL (see Supplementary Fig. 11 and movie 2). The image quality was lower than the Nikon camera, because there were fewer pixels and some compression in video mode, but it was sufficient. In the future, we are exploring this and other alternatives, such as coin microscopes, which are widely available for $20-60. Mini-centrifuges can be acquired for ~$50, although in the future, the centrifugation step could be skipped using swab squeeze tubes and tube filter caps. The remaining equipment components (flashlight, magnet and cuvette holder) are relatively small and inexpensive. We expect that these could be manufactured for <$5.
Regarding disposable costs, reagent use per test is very low. The primary disposable costs per test are as follows: Salivette saliva collection swab ($0.50), cuvette ($0.10), buoyant beads with antibody functionalization ($0.32), and magnetic beads with antibody functionalization ($0.15). This gives a cost-per-test of $1.07 at list prices. However, there are some minimum order costs, and we estimate that $6200 is enough to buy reagents and cuvettes for around 3600 tests, $1.72 per test (Supplementary Table 6). In practice, factors such as validation standards, storage and expiration, shipping, packaging, labor, and business expenses would increase total costs, while mass production and protocol optimization would help reduce them. Nonetheless, these values indicate that BAM tests have the potential to be affordable for most point-of-care applications.
This paper presents proof-of-principle for ultrasensitive BAM assays. To move towards real applications, we will need smaller and lighter instrumentation (e.g., using a coin camera or mobile phone). We must also simplify and automate the protocol to collect saliva, mix reagents, read results, and avoid accidental spillage. Reagent storage and shelf-life will require optimizing antibodies or antibody fragments, covalent attachment to the microspheres, and potentially lyophilization. Additionally, the experimental design can be optimized to achieve faster testing times and more effective signal responses. Parameters to optimize include: external magnet shape, position, field strength, orientation, field gradient, and magnetic separation time; sample volume; number of magnetic microspheres and buoyant microbubbles; microbubble diameter; antibody and microsphere functionalization; buffer optimization; sample collection/filtration protocols; instrument components and software optimization. Additionally, regulatory approval would be required on a finalized product prior to clinical use.
In conclusion, the BAM assay is designed to enhance antigen test sensitivity to qPCR-levels while remaining deployable. The method innovatively uses buoyant microbubbles, enabling rapid and efficient formation of BAM complexes with extremely low backgrounds. The BAM complexes scatter light intensely and rise, enabling counting with inexpensive portable cameras. Future work involves simplifying and optimizing the sample collection and analysis protocol for user-friendliness, developing integrated portable setups, automating the particle counting and analysis, testing in larger patient trials, and extending the approach to other disease-specific analytes.
Methods
Our method utilizes antibody-functionalized buoyant microbubbles and magnetic microspheres to capture and detect single molecules (Fig. 2A). This approach is based on research by McNaughton et al., employing BAM for the capture and separation of E. coli47. Our objective is to perform quantitative experiments on the analyte during the BAM complex separation stage by ensuring that the BAM complexes possess both magnetic and buoyant properties. In the initial step of this immunological analysis method, magnetic microspheres and buoyant microbubbles form sandwich antibody complexes. Assuming capture is stochastic and beads are roughly uniform in size at low concentrations, the number of molecules captured per microbubble follows a Poisson distribution29. When many microbeads are used to capture analytes at extremely low concentrations, only a small fraction of the microbeads are labeled, with the vast majority remaining unlabeled.
Materials
The magnet used in the BAM assay was made from a neodymium iron boride alloy with two shapes. The NdFeB magnet disc (catalog number D1051A) has a diameter of 8 mm, a thickness of 4 mm, and a Neodymium strength of 45. The magnet cylinder (cat. Cyl0164) has a diameter of 6 mm, a height of 8 mm, and a Neodymium strength of 50. They were both obtained from SuperMagnetMan (Pelham, AL, US). The LED light (Cat rechargeable extendable LED work light, cat. #: CT3115) was purchased from EZRED (Denville, NJ, US). The SARS-CoV-2 N-protein antibody, Rabbit Mab (Cat: 40143-R004 and Cat: 40143-R040) was obtained from Sino Biological (Wayne, PA, US). BSA powder (Bovine Serum Albumin, A7030-10G) was obtained from Sigma-Aldrich (St. Louis, MO, US). The semi-millimeter cuvette (product name: BRAND® semi-micro cuvette, Cat: BR759115) was purchased from MilliporeSigma (Burlington, MA, US). Low protein binding microcentrifuge tubes (Cat: 90401), Gibco PBS pH 7.2(Cat: 20012027) and the Pierce Antibody Biotinylation Kit for IP (Cat: 90407) were obtained from Thermo Scientific (Rockford, IL, US). A 66 mm width, 500 mm length optical rail (cat: XT66-500) and two 66 mm “Clamping Platform with Counterbored Slot” (Cat: XT66C4) were obtained from Thorlabs (Newton, NJ, US). Streptavidin Microbubbles (buoyant microbubbles) were purchased from Akadeum Life Sciences (Ann Arbor, MI, US). The magnetic beads were LodeStars Streptavidin 2.7 μm Magnetic Beads, which were provided by Agilent (Santa Clara, CA, US). The SARS-CoV-2 nucleocapsid protein (Cat: NUN-C5227-100ug, a 47.3 kDa protein synthesized with a his-tag) was obtained from Acro biosystems (Newark, DE, US). The biotin-mPEG 5 K (Cat: PLS-2054) was obtained from Creative PEGworks (Durham, NC, US). The Triton-X 100 and carboxymethylcellulose sodium salt were obtained from Alfa Aesar (Ward Hill, MA, US). All aqueous solutions used in the experiments were prepared using Milli-Q water (18.2 MΩ/cm) obtained from a ELGA Purelabflex2 system (ELGA LabWater, Woodridge, IL, US). The chemicals for lab-made, artificial saliva samples were as follows: Sodium chloride (NaCl) was obtained from Spectrum Chemicals (New Brunswick, NJ, US). Calcium chloride (CaCl2), magnesium chloride (MgCl2) and potassium phosphate dibasic (K2HPO4) were obtained from Sigma-Aldrich (St. Louis, MO, US). The carboxymethylcellulose sodium was obtained from Alfa Aesar (Ward Hill, MA, US). Potassium chloride (KCl) was obtained from Thermo Scientific (Rockford, IL, US). The urea was obtained from J.T. Baker (Phillipsburg, NJ, US).
Experimental setup
As shown in Fig. 6, the experimental setup consists of a 3D printed magnet slide holder, cuvette holder, six magnet discs, two magnet cylinders, a digital camera (Nikon D7000), two LED light sources, and a 66 mm optical rail (according to the materials section above). The upper end of the semi-milliliter cuvette was tilted 3° towards the camera direction and fixed onto the cuvette holder. The slight tilt angle prevents BAM complexes from touching or getting stuck on the cuvette walls as they rise. The setup includes six magnetic discs and two magnetic cylinders, with each set comprising three magnetic discs and one magnetic cylinder. Two magnetic discs are positioned at the bottom of the magnet slide holder, while one magnetic disc and one magnetic cylinder are placed on top. The magnetic field from the magnets was oriented normal to the cuvette bottom and towards the bottom front of the cuvette. The field and field gradient were sufficient to capture all magnetic and BAM complexes within 1 min and allow high capture efficiency, which was deemed sufficient for our purposes, and the configuration has not been further optimized. The magnets for both cuvettes are slid into place or removed together, synchronizing the attraction and release of the BAM complexes in both cuvettes. The LED lights placed on both sides of the cuvettes are turned on, and the camera position is adjusted on the optical rail until a clear image of the two cuvettes appears in the camera view.
A Schematic illustration of the instrument setup, including flashlights, a camera, a cuvette holder, and permanent magnets. B Photograph of the BAM test setup. The cuvettes are placed on the cuvette holder with magnets under each cuvette. The camera and cuvette holder are attached to an optical rail. Two flashlight sources illuminate the cuvettes from the side. C Photo showing the cuvette and magnets. D BAM assay tack analysis gray-map.
To study the effect of temperature within reasonable temperature ranges when environments/rooms are not heated or cooled, we performed the BAM assay at 5 °C, room temperature (18–20 °C) and 40 °C. For the tests conducted at 5 °C, we relocated all equipment to the controlled environment room (Supplementary Fig. 12A), manufactured by Climate Technologies, Inc. (Laytonsville, MD, US), model G3. For the tests at 40 °C, the equipment was placed in a metallic cabinet featuring a transparent acrylic door with hinges for opening and closing (Supplementary Fig. 12B). The side of the cabinet features a circular aperture with a diameter of 10 cm, designed for the introduction of heated air stream. This configuration allows the cabinet to be isolated during heating. Temperature control at 40 °C was achieved through an air steam stage incubator, model ASI400, manufactured by NEVTEK (Burnsville, VA, US).
Protocols
Antibody biotinylation
The antibodies immobilized on the microbubbles were SARS-CoV-2 N-protein antibody, Rabbit Mab (40143-R004, referred to as antibody (R004) below). The antibodies immobilized on the magnetic microspheres were SARS-CoV-2 N-protein antibody, Rabbit Mab (40143-R040, referred to as antibody (R040) below). Each of these antibodies binds to a different and non-overlapping epitope on the N-protein so that BAM complexes can form. Since there is only one of each epitope per protein, the analyte is not “lost” in buoyant-analyte-buoyant or magnetic-analyte-magnetic complexes. To biotinylate the N-protein antibodies (Ab-R004 and Ab-R040), the “Pierce Antibody Biotinylation Kit for IP” (Catalog #90407, Thermofisher Scientific, Waltham, MA, USA) was utilized, which reacts with amine groups on the antibodies through an NHS-PEG4-biotin linker. The antibody biotinylation kit includes a microtube containing solid NHS-PEG4-biotin, Zeba Spin desalting columns, and 10 mL of 20× PBS solution. Firstly, dilute the 20× PBS solution to 1× PBS (10 mM sodium phosphate, 0.15 M NaCl, pH 7.5) and add 100 μL of 1× PBS solution to the microtube containing solid NHS-PEG4-biotin to prepare an 8.5 mM solution. Modifying 50 μg IgG antibody was calculated to require 1.6 μL of 8.5 mM NHS-PEG4-biotin solution at a 40-fold biotin linker concentration. Then, 50 μL of 1 μg/μL Ab-R004 and Ab-R040 were separately placed into 2 mL low protein-binding microcentrifuge tubes, followed by the addition of 1.6 μL of 8.5 mM NHS-PEG4-biotin solution and 48.4 μL of 1× PBS solution. The mixture was incubated at room temperature for 30 min. Simultaneously, two Zeba Spin Desalting Columns were prepared by removing the bottom closure and loosening the top cap. The Zeba Spin Desalting Columns were placed in separate 2 mL microcentrifuge tubes and centrifuged at 1500 × g for 1 min, discarding the solution in the centrifuge tubes. This step was repeated twice by adding 300 μL of 1× PBS solution, centrifuging at 1500 × g for 1 min, and discarding the solution. Subsequently, the incubated antibody solutions were added individually onto the compact resin bed of each Desalting column, placed in a new low protein binding microcentrifuge tube, and centrifuged at 1500 × g for 2 min. At this point, the antibody concentration in the solution was 0.5 μg/μL. The biotinylated NC antibodies were then diluted to 0.05 μg/mL by 1× PBS solution and stored at −20 °C.
Surface modification of buoyant microbubbles and magnetic microspheres
100 μL of the streptavidin-coated buoyant microbubbles and 12 μL of the streptavidin-coated magnetic microspheres were added to separate low protein-binding microcentrifuge tubes to functionalize them with biotinylated antibody and biotinylated PEG. 2.6 μL of biotinylated-antibody (R004) was added to the buoyant microbubble solution, and 2.6 μL of biotinylated-antibody (R040) was added to the magnetic microsphere solution. The solution was pipetted rapidly up and down to mix. After antibody modification, 100 μL of 0.01 mM biotin-mPEG 5k was added to each centrifuge tube to block the empty streptavidin site, and the solution was pipetted rapidly up and down to mix. Then, 300 μL of 1× PBS was added, and the tubes were centrifuged at 400 × g for 3 min. The subnatant was aspirated from the buoyant microbubble solution, and the supernatant was aspirated from the magnetic microsphere solution. This washing process was repeated twice. After the washing steps, a 1× PBS solution containing 2% bovine serum albumin (BSA) and 1% Triton-X 100 were added, and the tubes were incubated overnight at 4 °C. The microbeads were washed thrice with 400 μL, 1× PBS under centrifugation 400 × g for 3 min. Finally, the buoyant microbubbles and magnetic microspheres were stored in 400 μL of a 1× PBS solution containing 0.5% BSA, 0.1% Triton-X 100, with a concentration of 2.5 × 104 particles/μL at 4 °C. These were used within 24 h of functionalization.
N-protein reconstitution
The lyophilized SARS-CoV-2 N-protein for the spiking saliva sample was reconstituted by 400 μL 1× PBS with a concentration of 250 μg/mL. It was then diluted to the desired concentration with 1× PBS solution. The final concentration of each bead was approximately 750,000 beads for each test, meaning 750,000 beads per 30 μL of solution.
BAM test protocol
30 μL of the antibody-modified buoyant microbubbles and 30 μL of the magnetic microspheres were transferred into a 2 mL centrifuge tube using a micropipette. 10 μL of the N-protein solution was subsequently added to the tube, and the mixture was incubated on a rotator at room temperature for 30 min. After incubation, 130 µL of 1× PBS was added to the mixture. The entire solution was then transferred from the centrifuge tube to a semi-milliliter cuvette, which was placed into the 3D-printed cuvette holder. The magnet was placed under the cuvette for 15 min to collect BAM complexes to pull down the magnetic microspheres while unbound microbubbles floated upward. The magnet was then removed, and the trajectories of the BAM complexes were monitored through time-lapse photography. We connected the Nikon D7000 SLR digital camera to the laptop using a USB cable and controlled the camera through the Nikon Camera Control Pro 2 version 2.35.1 software. The D7000 camera has a 4928 x 3264 pixel detector, and was focused on the front of the two cuvettes, so there were ~195 pixels/mm. The camera settings were configured with a shutter speed of 1/1.3 s, an aperture of f/40, and ISO 250. In the Interval Time Shooting mode, we set the total number of shots to 310 with a 2-s interval. The choice of 310 shots allowed us 20 s to remove the magnet from the cuvette holder. Subsequently, all the photos were transferred to Microsoft’s Video Editor software, where the 300 photos were transformed into a 10-s video with a frame rate of 30 FPS. A MATLAB script was developed to visualize the trajectories of the BAM complexes through every frame of the video (Supplementary Figs. 10, 13, 14 and Table 5 for the code and processing results).
To show that BAM assay could be performed with a cell phone camera, we replaced the Nikon camera with a Samsung Galaxy Z Flip6 phone. The phone was folded at a right angle and set on its side on a flat surface (where it could stand without additional support). The phone camera was focused on the cuvette as shown in Supplementary Fig. 11. The phone was set to lock focus, 2.4× zoom, disable auto beautify and filters, and enable HEVC mode. The phone camera recorded a real-time video with a resolution of 1280 × 720, there were ~40 pixels/mm.
Human saliva collection protocol
The saliva collection protocols were approved by the Institutional Review Boards (IRB) of Clemson University (IRB2021-0703) and Prisma Health (Pro00100731). Experiments with the saliva were performed under approved Clemson University institutional biosafety committee (IBC) protocol IBC2022-0138. All study participants provided their written informed consent to provide a saliva sample. Participants provided 1–2 mL of saliva collected in a 50 mL conical tube and some demographic data (age, gender, race/ethnicity) to the research study48. Samples were deidentified and stripped of personal identifying information before being passed to the research team. Part of the samples (200 ml) was used for a RT-PCR clinical diagnostic test49,50 and eventual sequencing for SARS-CoV2 variant identification51,52.
Patient saliva samples treatment
The centrifuge tubes were thawed and opened within a glove bag (No. 690323) from NPS (Green Bay, WI, US) in a biosafety hood (Class II type A2) from Labconco (Kansas, MO, US). To lyse the viruses in saliva and release the encapsulated N-proteins, 400 μL of saliva sample was mixed with 400 μL of 1% Triton-X 1× PBS solution by pipette in the glove bag. Welch et al. report 5.9 logs reduction in 2 min with 0.5% Triton-X53. We applied the mixture to a Salivette cotton swab Salivette (REF: 51.1534 from Sarstedt AG, Nümbrecht, Germany) and expressed the fluid by centrifuging in the Salivette kit centrifuge tube at 1000 × g for 2 min to filter and remove any debris. The saliva supernatant was collected for testing. Processed saliva samples stored at freezing temperatures may experience flocculation upon thawing. The samples were centrifuge at 400 × g for 5 min to collect the supernatant for BAM testing.
Dilution of patient saliva supernatant
The supernatant was transferred to a low protein binding centrifuge tube, where it was diluted with a 0.5% Triton-X 100, 1× PBS solution. In each subsequent serial dilution step, the solution was diluted tenfold, with replacement of the pipette tip after each dilution.
Simulated saliva studies
The artificial saliva used in these studies was artificially made using the following: 15.6 mM NaCl, 16.5 mM KCl, 1.01 mM CaCl2, 0.361 mM MgCl2, 2.07 mM K2HPO4, 16.3 mM urea, and 5.0 g/L carboxymethylcellulose sodium salt in one aqueous liter54. 10 μL of 10% simulated saliva (1 × PBS) was spiked with N-protein. Each BAM test was run per the protocol discussed in the section “The BAM Test Protocol.” Final concentrations of N-protein in the 10 μL of 10% simulated saliva varied between 0 and 12 fg/mL, with each 1 fg/mL corresponding to 127 N-proteins. In the low-concentration samples (0–2 fg/mL), relatively few BAM complexes were seen, which could be easily counted. However, in the more concentrated samples (6 and 12 fg/mL), the large number of BAM complexes resulted in overlapping tracks, which made it difficult to count. To combat this, the samples were thoroughly mixed, and one-tenth of their total volume was taken as a representative sample to estimate the total number of BAM complexes in the entire sample.
Spiked negative patient saliva assays
PCR-negative patient specimens were collected by thawing the stored frozen specimens and pipetting them onto a Salivette swab. To remove debris, the swab was then placed into a centrifuge (provided in the purchased Salivette swab kit). Apart from centrifugation, squeezing the swab with a syringe also works. Without this step, it was found that the number of nonspecifically bound BAM complexes rose significantly. These steps mimic either future direct Salivette collection or integrated filtration steps. Each BAM test was run per the protocol discussed in the section “The BAM Test Protocol.”
Positive and negative patient studies
These studies were conducted on n = 3 PCR-positive and n = 8 PCR-negative saliva specimens. Because our setup allows for two samples to be tested simultaneously, a PCR-positive specimen (left cuvette) was run alongside a PCR-negative specimen (right cuvette) (Fig. 4A and Supplementary Movie 1). Each BAM test was run per the protocol discussed in the section “The BAM Test Protocol.”
Serial dilution and quantification studies
A qualitative positive-or-negative result is sufficient for most uses of rapid infection assays. Even when the test provides quantification (e.g., Ct value in qPCR), thresholds are usually applied to determine infection status, and patients are usually given the same treatment regardless of the Ct value, especially in initial diagnosis. Nonetheless, analyte quantification is useful for some applications, and we were concerned that the number of BAM complexes in the patient samples would not remain linear through all ranges when compared to the PCR Ct values. To test this, an additional 10 μL of 6 ng/mL N-protein was spiked into the patient saliva samples. Their fit to the expected linear regression was then analyzed.
Given the difficulty of counting more than ~200 BAM complexes due to overlapping tracks and non-linearities due to aggregation and saturation, quantification requires diluting the specimen until it is in a linear range. We thus acquired n = 8 PCR-positive specimens, with Ct values ranging between 18 and 36 (2.5 × 104 −0.7 cp/µL according to prior calibrations) and performed a BAM assay on each specimen. If the test was positive, the specimen was serially diluted by 10 or 100× and tested again until diluted to a point where the BAM counts were in the linear range, which was between the LOD (61 counts) and ~200 counts. This required up to 106-fold dilution for the concentrated specimens. After dilution to the linear range, the samples were tested in triplicate, and their results were compared to the RNA concentrations calculated based on the sample’s Ct value. After these analyses, the specimens were further diluted by a factor of 100× (up to 108-fold overall) to see the background with almost no analyte. The concentration for each trial was quantified by subtracting the average background signal (20 BAM complexes) from the BAM counts in the linear region and then dividing by the dilution factor. This method is conceptually similar to quantitative PCR, where DNA concentration is determined based on when the fluorescence signal crosses and remains above a threshold. In our case, however, we serially dilute the specimen until the BAM signal falls below the threshold and stays there.
Figure creation
All figures are original, and were created using the following software: PowerPoint (Figs. 1, 2, 4, 6, ToC Figure, and Supplementary Figs. 5, 6, 8, and 12), Excel (Figs. 3 and 5A), MATLAB (Fig. 4A and Supplementary Figs. 1, 2, 4–6, 8, 10, 11, and 13), and Origin (Figs. 4B, 5B, and Supplementary Figs. 3, 7, and 9).
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files or are available from the corresponding author upon reasonable request.
Code availability
The MATLAB code used for this study is available in the Supplementary Fig. 14. The software utilized is MATLAB R2023a.
References
Msemburi, W. et al. The WHO estimates of excess mortality associated with the COVID-19 pandemic. Nature 613, 130–137 (2023).
Danlos, F. X. et al. Metabolomic analyses of COVID-19 patients unravel stage-dependent and prognostic biomarkers. Cell Death Dis. 12, 258 (2021).
Kermali, M., Khalsa, R. K., Pillai, K., Ismail, Z. & Harky, A. The role of biomarkers in diagnosis of COVID-19—a systematic review. Life Sci. 254, 117788 (2020).
Ponti, G., Maccaferri, M., Ruini, C., Tomasi, A. & Ozben, T. Biomarkers associated with COVID-19 disease progression. Crit. Rev. Clin. Lab. Sci. 57, 389–399 (2020).
Hunt, P. W., Lee, S. A. & Siedner, M. J. Immunologic biomarkers, morbidity, and mortality in treated HIV infection. J. Infect. Dis. 214, S44–50 (2016).
Land, K. J., Boeras, D. I., Chen, X. S., Ramsay, A. R. & Peeling, R. W. REASSURED diagnostics to inform disease control strategies, strengthen health systems and improve patient outcomes. Nat. Microbiol. 4, 46–54 (2019).
Kettler, H., White, K. & Hawkes, S. J. Mapping the Landscape of Diagnostics for Sexually Transmitted Infections: Key Findings and Recommendations (World Health Organization, 2004).
World Health Organization. Target Product Profiles for Priority Diagnostics to Support Response to the COVID-19 Pandemic v. 1.0. (World Health Organization, 2020).
Klein, D. Quantification using real-time PCR technology: applications and limitations. Trends Mol. Med. 8, 257–260 (2002).
Park, J. S., Kim, S., Han, J., Kim, J. H. & Park, K. S. Equipment-free, salt-mediated immobilization of nucleic acids for nucleic acid lateral flow assays. Sens. Actuators B Chem. 351, 130975 (2022).
Budd, J. et al. Lateral flow test engineering and lessons learned from COVID-19. Nat. Rev. Bioeng. 1, 13–31 (2023).
Kortela, E. et al. Real-life clinical sensitivity of SARS-CoV-2 RT-PCR test in symptomatic patients. PLoS ONE 16, e0251661 (2021).
Stanley, S. et al. Limit of detection for rapid antigen testing of the SARS-CoV-2 Omicron and Delta variants of concern using live-virus culture. J. Clin. Microbiol. 60, e0014022 (2022).
Mak, G. C. et al. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J. Clin. Virol. 129, 104500 (2020).
Krenn, F. et al. Ten rapid antigen tests for SARS-CoV-2 widely differ in their ability to detect Omicron-BA.4 and -BA.5. Med. Microbiol. Immunol. 212, 323–337 (2023).
Koczula, K. M. & Gallotta, A. Lateral flow assays. Essays Biochem. 60, 111–120 (2016).
Winnett, A. et al. SARS-CoV-2 viral load in saliva rises gradually and to moderate levels in some humans. medRxiv, https://doi.org/10.1101/2020.12.09.20239467 (2020).
Liu, Z. et al. Simultaneously ultrasensitive and quantitative detection of influenza A virus, SARS-CoV-2, and respiratory syncytial virus via multichannel magnetic SERS-based lateral flow immunoassay. Nanomed. Nanotechnol., Biol. Med. 47, 102624 (2023).
González-Cabaleiro, L. et al. Pushing the limits of lateral flow immunoassay by digital SERS for the ultralow detection of SARS-CoV-2 virus. Small Sci. 4, 2400259 (2024).
Guo, K. et al. Rapid single-molecule detection of COVID-19 and MERS antigens via nanobody-functionalized organic electrochemical transistors. Nat. Biomed. Eng. 5, 666–677 (2021).
Hellewell, J. et al. Feasibility of controlling COVID-19 outbreaks by isolation of cases and contacts. Lancet Glob. Health 8, e488–e496 (2020).
Kucharski, A. J. et al. Effectiveness of isolation, testing, contact tracing, and physical distancing on reducing transmission of SARS-CoV-2 in different settings: a mathematical modelling study. Lancet Infect. Dis. 20, 1151–1160 (2020).
MacIntyre, C. R. Case isolation, contact tracing, and physical distancing are pillars of COVID-19 pandemic control, not optional choices. Lancet Infect. Dis. 20, 1105–1106 (2020).
Fan, W., Dong, Y., Ren, W. & Liu, C. Single microentity analysis-based ultrasensitive bioassays: recent advances, applications, and perspectives. TrAC Trends Anal. Chem. 162, 117035 (2023).
Sheehan, P. E. & Whitman, L. J. Detection limits for nanoscale biosensors. Nano Lett. 5, 803–807 (2005).
Tamanaha, C. R., Mulvaney, S. P., Rife, J. C. & Whitman, L. J. Magnetic labeling, detection, and system integration. Biosens. Bioelectron. 24, 1–13 (2008).
Tsai, H., Lin, Y., Chang, H. W. & Fuh, C. B. Integrating the QCM detection with magnetic separation for on-line analysis. Biosens. Bioelectron. 24, 485–488 (2008).
Rissin, D. M. et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat. Biotechnol. 28, 595–599 (2010).
Gorris, H. H. & Walt, D. R. Mechanistic aspects of horseradish peroxidase elucidated through single-molecule studies. J. Am. Chem. Soc. 131, 6277–6282 (2009).
Nam, J. M., Thaxton, C. S. & Mirkin, C. A. Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science 301, 1884–1886 (2003).
Taylor, S. W., Wang, C. & Anker, J. N., https://doi.org/10.26434/chemrxiv-2024-501wj (2024).
Shi, G. et al. Isolation of rare tumor cells from blood cells with buoyant immuno-microbubbles. PLoS ONE 8, e58017 (2013).
Chan, K. Y. et al. Ultrasensitive detection of E. coli O157:H7 with biofunctional magnetic bead concentration via nanoporous membrane based electrochemical immunosensor. Biosens. Bioelectron. 41, 532–537 (2013).
Baldrich, E. & Munoz, F. X. Carbon nanotube wiring: a tool for straightforward electrochemical biosensing at magnetic particles. Anal. Chem. 83, 9244–9250 (2011).
Chorti, P. & Christodouleas, D. C. Sink/Float magnetic immunoassays for in-field bioassays. Angew. Chem. Int Ed. Engl. 60, 26947–26953 (2021).
Shan, D. et al. N-protein presents early in blood, dried blood and saliva during asymptomatic and symptomatic SARS-CoV-2 infection. Nat. Commun. 12, 1931 (2021).
Park, J. et al. An integrated magneto-electrochemical device for the rapid profiling of tumour extracellular vesicles from blood plasma. Nat. Biomed. Eng. 5, 678–689 (2021).
Berg, H. C. & Purcell, E. M. Physics of chemoreception. Biophys. J. 20, 193–219 (1977).
Gomez, S. M., Choy, G., Kabir, N. & Leonard, E. F. Capture of rare cells in suspension with antibody-coated polystyrene beads. Biotechnol. Prog. 15, 238–244 (1999).
Guasto, J. S., Rusconi, R. & Stocker, R. Fluid mechanics of planktonic microorganisms. Annu. Rev. Fluid Mech. 44, 373–400 (2012).
Kamat, V., Rafique, A., Huang, T., Olsen, O. & Olson, W. The impact of different human IgG capture molecules on the kinetics analysis of antibody-antigen interaction. Anal. Biochem. 593, 113580 (2020).
Landry, J. P., Ke, Y., Yu, G. L. & Zhu, X. D. Measuring affinity constants of 1450 monoclonal antibodies to peptide targets with a microarray-based label-free assay platform. J. Immunol. Methods 417, 86–96 (2015).
Marin, L. M. et al. Identification of SARS-CoV-2 biomarkers in saliva by transcriptomic and proteomics analysis. Clin. Proteom. 20, 30 (2023).
Zhao, H. et al. Assembly of SARS-CoV-2 nucleocapsid protein with nucleic acid. Nucleic Acids Res. 52, 6647–6661 (2024).
Paegel, B. M., Grover, W. H., Skelley, A. M., Mathies, R. A. & Joyce, G. F. Microfluidic serial dilution circuit. Anal. Chem. 78, 7522–7527 (2006).
Schmidt, C. et al. A multiparametric fluorescence assay for screening aptamer-protein interactions based on microbeads. Sci. Rep. 12, 2961 (2022).
McNaughton, B. H., Anker, J. N. & Kinnunen, P. K. Buoyant-antigen-magnetic (BAM) immunoseparation, isolation, and detection of specific pathogenic bacterial cells. 2022.2011. 2027.517249, https://doi.org/10.1101/2022.11.27.517249 (2022).
King, K. L. et al. Repurposing a SARS-CoV-2 surveillance program for infectious respiratory diseases in a university setting. Front. Public Health 11, 1168551 (2023).
Vogels, C. B. F. et al. SalivaDirect: a simplified and flexible platform to enhance SARS-CoV-2 testing capacity. Med 2, 263–280.e266 (2021).
Ham, R. E. et al. Efficient SARS-CoV-2 quantitative reverse transcriptase PCR saliva diagnostic strategy utilizing open-source pipetting robots. J. Vis. Exp. https://doi.org/10.3791/63395 (2022).
Ham, R. E. et al. Identifying SARS-CoV-2 variants of concern through saliva-based RT-qPCR by targeting recurrent mutation sites. Microbiol. Spectr. 10, e0079722 (2022).
King, K. L. et al. SARS-CoV-2 variants of concern Alpha and Delta show increased viral load in saliva. PLoS ONE 17, e0267750 (2022).
Welch, S. R. et al. Analysis of inactivation of SARS-CoV-2 by specimen transport media, nucleic acid extraction reagents, detergents, and fixatives. J. Clin. Microbiol. 58, https://doi.org/10.1128/JCM.01713-20 (2020).
Darvell, B. W. The development of an artificial saliva for in vitro amalgam corrosion studies. J. Oral. Rehabil. 5, 41–49 (1978).
Acknowledgements
This research was financially supported by “Buoyant and Magnetic (BAM) Assays for On Site Sensitive Rapid Diagnostics,” COVID Testing Research Seed Grant Program from Clemson University and “Rising to the Separations and Diagnostic Challenge with Buoyancy and Magnetism (BAM),” Agilent Technologies Inc., Applications and Core Technology—University Relations (ACT-UR), and the Clemson University Creative Inquiry and Summer UR programs.
Author information
Authors and Affiliations
Contributions
The authors had the following CRediT taxonomy contributor roles: C.W.: Investigation, methodology, software, visualization, writing—original draft, and writing—review and editing; E.S.: investigation, validation, writing review and editing; C.B.L.: funding acquisition, writing—review and editing; D.D.: methodology, investigation, funding acquisition, resources, validation, writing—review and editing; J.N.A.: conceptualization, resources, formal analysis, methodology, project administration, software, supervision, visualization, funding acquisition, writing—original draft, and writing—review and editing.
Corresponding author
Ethics declarations
Competing interests
J.N.A. and C.B.L. are Scientific Advisors for Akadeum Life Sciences, the company that sells the buoyant microbubbles used in the BAM assay, and which owns the rights to a patent describing buoyant and magnetic analyte labeling and detection (US Patent 10,724,930). All other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, C., Satterfield, E., Livi, C.B. et al. Rising to the ultrasensitive rapid diagnostic challenge with buoyant-analyte-magnetic (BAM) assays. npj Biosensing 2, 34 (2025). https://doi.org/10.1038/s44328-025-00054-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s44328-025-00054-x