Abstract
Hepatocellular carcinoma (HCC) is the most common primary liver cancer and a major cause of cancer-related deaths worldwide. Conventional diagnostics lack sensitivity for early detection. This review summarizes emerging biomarkers, advanced molecular classifications, and technologies such as RNA sequencing and liquid biopsy that enhance tumor characterization and monitoring. While promising, these approaches face validation and accessibility barriers. Their integration into clinical practice could enable personalized therapy and improve outcomes.
Similar content being viewed by others
Introduction
Hepatocellular carcinoma (HCC) remains one of the leading causes of cancer-related mortality worldwide, reflecting the limitations of current diagnostic and therapeutic strategies. Despite advancements in surveillance, traditional diagnostic tools such as ultrasound and serum markers like alpha-fetoprotein (AFP) lack sufficient sensitivity and specificity for early detection, resulting in late-stage diagnoses and poor clinical outcomes. Consequently, there is an urgent need to explore novel biomarkers and advanced molecular technologies to improve the early diagnosis and prognostic assessment of HCC.
In this review, we will discuss the latest advancements in biomarker discovery for HCC, focusing on innovative candidates that have demonstrated potential for clinical application. Particular attention will be given to novel molecular markers such as DCLK1, GPC3, CD276, and OPN, which have shown promising results in identifying early-stage HCC and predicting disease progression. Additionally, we will explore cutting-edge technologies like single-cell sequencing and liquid biopsy, which are revolutionizing our understanding of HCC biology and enabling more precise patient stratification.
Finally, we will address how these emerging biomarkers and technological innovations are contributing to novel molecular classification systems for HCC. By integrating these new discoveries with genetic and immunological profiling, it is now possible to redefine HCC subtypes with greater accuracy, paving the way for personalized therapeutic strategies. This review aims to provide a comprehensive overview of these advancements, highlighting their implications for clinical practice and future research directions.
HCC: epidemiology and diagnostic strategies applied in current clinical practice
Hepatocellular carcinoma (HCC) is the sixth most common neoplasia worldwide and the most common form of primary liver cancer, accounting for approximately 75–85% of cases. Originating from hepatocytes, the liver’s main cells, HCC typically develops in the context of chronic liver disease, often associated with cirrhosis. Its incidence varies significantly across different geographic regions, reflecting the distribution of risk factors. Furthermore, its global incidence is on the rise, making it a major cause of cancer-related mortality worldwide1,2,3,4.
Numerous risk factors have been identified for HCC. Chronic infection with hepatitis B virus (HBV) and hepatitis C virus (HCV) is are main contributor, particularly in regions such as East Asia and sub-Saharan Africa for HBV, and Western Europe and North America for HCV1,5. Alcohol abuse, leading to fatty liver disease and cirrhosis, is another significant determinant. In recent decades, metabolic dysfunction-associated steatotic liver disease (MASLD), previously named non-alcoholic fatty liver disease (NAFLD), has emerged as the primary cause of chronic liver disease globally, reflecting global changes in lifestyle and dietary habits1,6. MASLD is also the primary driver of the rising incidence of HCC, with the annual occurrence of MASLD-related HCC projected to increase by 45–130% by 20307.
The highest risk of HCC is observed in patients with advanced fibrosis or cirrhosis. However, numerous studies have shown that 20–50% of HCC cases occur in individuals with MASLD, even in the absence of cirrhosis8. Finally, other risk factors for HCC include exposure to aflatoxins, carcinogenic substances produced by molds in poorly stored foods, and genetic conditions such as hereditary hemochromatosis1,5. The management of HCC is complex and requires a multidisciplinary approach. Major international guidelines, including those from the European Association for the Study of the Liver (EASL)1 and the American Association for the Study of Liver Diseases (AASLD)4, emphasize the importance of a regular biannual screening in high-risk patients, such as those with cirrhosis, using ultrasound and alpha-fetoprotein measurements. Abdominal Ultrasound (US) is the most commonly used surveillance tool for HCC due to its accessibility, low cost, and non-invasiveness. It is recommended as the first-line tool for its ability to detect liver nodules. However, the effectiveness of ultrasound is influenced by factors such as obesity and the presence of severe steatosis, which can limit adequate visualization of liver parenchyma. Additionally, the sensitivity of ultrasound depends on the operator’s experience, which can lead to variability in results1,4.
Magnetic Resonance Imaging (MRI) and Computed Tomography (CT) are used to confirm the nature of suspicious lesions identified during ultrasound and to complete tumor staging. These exams are more expensive and require the use of contrast agents, which may not be suitable for patients with severe renal impairment1,4. Abbreviated MRI has been proposed as an alternative also for screening, but there are limited data to support its use, even when restricted to some specific subsets of patients9. Although less commonly used than MRI e CT, contrast-enhanced ultrasound (CEUS) represents a valuable diagnostic tool for HCC, particularly for patients for whom CT or MRI are not feasible. Its utility is largely attributed to the exclusive intravascular distribution of the contrast agent, which allows for real-time assessment of vascular dynamics and of HCC characteristic enhancement patterns10.
Another HCC diagnostic tool is represented by tumor markers, such as alpha-fetoprotein (AFP). Although AFP is still widely used as a screening marker for HCC, its sensitivity and specificity are insufficient. Only about 50% of patients with HCC present elevated AFP levels, since some HCCs, particularly in the early stages, may not produce AFP at all11,12. On the other hand, AFP concentrations can be elevated in the absence of HCC, such as in patients with active hepatitis. Despite this, current guidelines suggest the combined use of US and AFP in HCC screening, since this approach translates into greater sensitivity with an acceptable reduction in specificity1. Notably, in the last EASL guideline, the use of lens culinaris agglutinin-reactive alpha-fetoprotein (AFP-L3), angiopoietin-2 (ANG2), and the combination of AFP with Des-gamma-carboxyprothrombin (DCP), has been proposed to improve the performance of AFP, but these new markers have significant limitations. AFP-L3 is a specific fraction of AFP more closely associated with HCC, but it is not sensitive enough to be used as a sole indicator. ANG2, a marker for angiogenesis, was proposed as a serum tumor biomarker despite it is less studied. DCP is a marker produced by tumor cells in HCC, but it can also be elevated in other non-tumor conditions, limiting its clinical utility11,12. Moreover, composite biomarker models, such as the GALAD score, have been developed. This score is the most thoroughly validated integrative tool, combining gender, age, AFP, AFP-L3, and DCP, demonstrating 82% sensitivity and 89% specificity, with an AUROC of 0.92 for HCC detection. Notably, even in early-stage HCC, it maintained acceptable performance (sensitivity 73%, specificity 87%)13. More recently, novel investigational models have emerged, such as the HCC Early Detection Screening (HES) score and the aMAP score. The original HES score combined AFP with age, alanine aminotransferase, and platelet count14. The updated HES v2.0, which incorporates AFP-L3 and DCP, has shown a 6–15% higher sensitivity than GALAD during 1–2 years of surveillance15. Finally, aMAP score, which integrates age, sex, albumin-bilirubin, and platelet count, is currently under investigation as a risk stratification tool for personalized HCC surveillance in patients with chronic liver disease16.
Histological features of HCC and molecular correlates
Morphologically, HCC is characterized by hepatocytic differentiation, which can be identified through routine microscopy and supported by immunohistochemical and molecular analyses. HCC disrupts the normal hepatic architecture, primarily through the absence of portal tracts and the distortion or loss of the reticulin framework17. These architectural changes result from tumor expansion and the replacement of normal hepatic parenchyma with malignant cells. HCC demonstrates four principal histological growth patterns. First, the trabecular pattern, the most common growth pattern, is characterized by malignant hepatocytes arranged in broad plates or cords, usually more than three cells thick. These trabeculae are separated by sinusoids, which may show endothelial wrapping or transgressing vessels18. Second, the solid pattern, composed of densely packed tumor cells with minimal intervening stroma or sinusoidal spaces. This pattern is frequently associated with high-grade tumors and poor differentiation18. Third, the pseudo-glandular (pseudo-acinar) pattern, in which tumor cells form gland-like structures resembling acini, which may contain bile or necrotic debris. This pattern highlights the ability of malignant hepatocytes to mimic glandular differentiation18. Fourth, the macrotrabecular pattern, characterized by thick trabeculae ( ≥ 10 cells), is often associated with an aggressive clinical course and worse prognosis18. Mixed patterns are observed in about 50% of cases, emphasizing the histological heterogeneity of HCC. This heterogeneity can reflect tumor progression or dedifferentiation.
The tumor often shows increased arterialization, characterized by the presence of abnormal arterioles within the parenchyma and sinusoidal capillarization. This vascular transformation is a hallmark of HCC and reflects the dependency of the tumor on arterial blood supply. Sinusoids within the tumor show capillarization, with endothelial cells expressing markers like CD34, reflecting a shift from normal hepatic to tumor-associated vasculature. Small aggregates of macrophages or peliosis-like areas may also be present within the tumor sinusoids, further highlighting its complex microenvironment19.
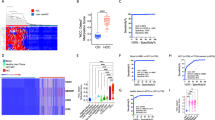
The World Health Organization (WHO) 2019 classification recognizes several histological subtypes of HCC, each with distinct clinical and molecular features20. In their review, Choi and Thung described very well all HCC subtypes divided for histological and mutational features20. These subtypes include fibrolamellar carcinoma, macrotrabecular massive HCC, scirrhous HCC, clear cell HCC, and chromophobe HCC, among others; the full list is available in Table 1, and in Fig. 1 shows histological features of some HCC subtypes. A close connection exists between HCC subtypes and specific genetic mutations, which influence their clinical behavior and prognosis. Fibrolamellar carcinoma, for instance, occurs predominantly in younger patients without underlying liver disease and is characterized by eosinophilic tumor cells and dense intratumoral fibrosis with a lamellar pattern. Molecularly, it involves the DNAJB1::PRKACA fusion gene. Scirrhous HCC, characterized by abundant fibrosis, is often associated with mutations in the TSC1/2 genes and activation epithelial-to-mesenchymal transition signaling pathway. Similarly, chromophobe HCC is linked to the alternative lengthening of telomeres (ALT) mechanism, which allows telomere maintenance independent of telomerase reverse transcriptase (TERT) promoter mutations. Macrotrabecular massive HCC, defined by a growth pattern of thick trabeculae (≥10 cells), is linked to mutations in tumor protein p53 (TP53) and fibroblast growth factor (FGF) 19 amplifications correlating with its aggressive clinical course and high vascular invasion rates, often signifying a poor prognosis. In contrast, clear cell HCC, enriched in glycogen, exhibits fewer genetic alterations, contributing to its relatively favorable prognosis compared to other subtypes. Notably, immunohistochemistry (IHC) plays a pivotal role in diagnosing HCC and its subtypes. Markers such as arginase-1, Hep Par-1, and glypican-3 assist in identifying hepatocytic differentiation. Subtypes like scirrhous HCC, characterized by dense fibrosis, may express cytokeratin (CK) 7 and CK19, markers usually associated with cholangiocarcinomas.
The most common HCC subtypes are represented (courtesy of Prof. Simone Carotti and Dr. Andrea Baiocchini). All images are taken with NanoZoomer digital scanner, magnification 40X, linear bar 50 µm. Created with BioRender.com (https://BioRender.com/3fnrku2).
The histological and molecular diversity of HCC underscores the importance of comprehensive diagnostic approaches. Mixed histological patterns, seen in about half of HCC cases, reflect tumor progression and heterogeneity. Molecular alterations often accumulate during this progression, highlighting the dynamic nature of HCC. Understanding these changes is vital for advancing treatment strategies, including precision medicine.
However, we currently lack effective tools for the diagnosis and prognosis of early-stage HCC. For this reason, it is necessary to explore new biomarkers or to gain a better understanding of, and perhaps use differently, the current clinical biomarkers described in the guidelines.
New putative clinical biomarkers for an early diagnosis of HCC
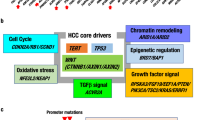
As mentioned above, current biomarkers and assays used in clinical practice are insufficient both to discriminate patients with HCC from those with pre-tumoral liver disease and to detect early HCC recurrence after surgical or medical treatment. For this reason, in recent years, several new biomarkers have been proposed with the aim to improve HCC prediction, screening, or monitoring. In this chapter, we will focus on new putative HCC biomarkers derived in recent years from studies on animal models and/or patient cohorts. (Table 2 and Fig. 2).
Schematic representation of proposed biomarkers for HCC diagnosis and prognosis, as summarized in the present review. Created with BioRender.com (https://BioRender.com/5xqg3hc).
Doublecortin-like kinase 1
Doublecortin-like kinase 1 (DCLK1) is a protein associated with microtubules in cytoplasm, involved principally in the regulation of microtubule polymerization and neurogenesis21,22. Several research teams described the overexpression of DCLK1 in different solid tumor type23,24,25,26,27,28,29. A strong relationship has been demonstrated between overexpression of DCLK1 and several tumor processes including tumor growth, metastasis, epithelial-to-mesenchymal transition (EMT), cancer stem cells (CSCs) self-renewal, tumoral microenvironment regulation21,22,30,31,32. Moreover, high DCLK1 is observed in patients with gastrointestinal tumors that have a worse prognosis33,34,35,36. Previous studies have linked increased DCLK1 level expression with poor prognosis in HCC, suggesting its potential as a biomarker for diagnosis and monitoring of disease progression. It was demonstrated that DCLK1 is targeted by microRNAs (miRNA) like miR-144 and miR-200a, which suppress DCLK1 expression, inhibiting cancer cell growth, stemness, and invasion. On the other hand, DCLK1 knockdown upregulates tumor suppressor miRNAs such as miR-143 and miR-145, reducing cell proliferation and migration37. Moore et al. have identified that miR-1246 and miR-184 were upregulated, while miR-206 was downregulated, in patients with high DCLK1 sera levels. Authors suggested that elevated DCLK1 and miR-1246 levels are associated with inflammation-driven tumorigenesis, while miR-206 reduction facilitates the transition from cirrhosis to HCC, and this could be used in clinical practice for patients suffering of chronic liver disease as biomarkers for following the progression from pre-neoplastic conditions to HCC and/or offer therapeutical targets37. Recent research associates DCLK1 with immune cell infiltration in cancers like gastric and colon cancer, particularly with tumor-associated macrophages (TAMs) and regulatory T cells (Tregs)33,38,39. Velazquez-Enriquez et al. have demonstrated that high DCLK1 expression in HCC patients correlates with increased infiltration of immune cells such as B cells, CD8 + T cells, CD4 + T cells, macrophages, neutrophils, dendritic cells, and myeloid-derived suppressor cells (MDSCs). Furthermore, DCLK1 shows significant correlations with Treg markers, including FOXP3, CCR8, STAT5B, and TGFB1, suggesting a role in promoting an immunosuppressive tumor microenvironment. Authors concluded that this interaction could help tumors evade immune surveillance, highlighting DCLK1 as a crucial regulator in immune suppression and a potential target for therapeutic intervention22. The possibility of monitoring HCC onset and progression through a simple serum analysis is very appealing. However, the mechanisms and functional role of DCLK1 need to be investigated more thoroughly before assuming a clinical role in liver cancer.
Glypican 3
Glypican 3 (GPC3) is a heparan sulfate proteoglycan that is rarely expressed in normal tissues; however, it is overexpressed in HCC and plays a role in HCC development40,41,42. In fact, GPC3 is overexpressed during the aggressive progression of HCC, indicating that it may play a role in the development of HCC41. Based on this, An et al. 40 have used anti-GPC3 nano-antibodies in immune-positron emission tomography (immunoPET) imaging to increase the diagnostic sensitivity for HCC, obtaining promising results in HCC animal models. However, tracers derived from nano-antibodies have the problem of being widely absorbed by the kidneys and this could compromise their clinical translation40. Another method to use the different expression of GPC3 was described by Cao et al. They developed two glypican-3 (GPC3)-specific CAR-NK-92 cell lines (GPC3-CAR-NK), GC33-G2D-NK and GC33-CD28-NK, and analyzed their effect in vitro and in vivo when used in combination with microwave ablation (MWA)42. Between the two kinds of engineered cells, GC33-G2D-NK showed a better degranulation upon stimulation and synergic antitumor effect with MWA against HCC42. The strength of this approach lies in the fact that MWA is already used in clinical practice, and clinical trials have been conducted using GPC3-specific CAR-NK cells43. The limitation of the study by Cao et al. was the use of NSG mice, since they lack a functional immune system, therefore preventing the interaction between CAR-NK cells and innate immune cells42. Li et al.41 performed a retrospective analysis based on GPC3 expression, demonstrating that MRI, radiomics, tumor morphology, and microvascular invasion (MVI) can noninvasively predict GPC3 expression in HCC patients41. Authors suggested integrating these indicators with clinical factors into nomograms to offer valuable insights for tailoring personalized treatment plans for patients diagnosed with HCC prior to surgery41.
The ability to both detect the presence of HCC and use specific NK cells to eliminate cancer cells makes GPC3 a promising clinical marker. However, further studies are needed before GPC3-oriented CAR-NK cells can be used in clinical practice.
CD 276
Cluster of differentiation (CD) 276, or B7-H3, is the principal member of B7 superfamily. PD-L1 (B7-H1) is another member of this superfamily5,44. In normal human tissues, CD276 is broadly expressed at the mRNA level but its protein expression is typically limited, suggesting post-transcriptional regulation. At the same time, abnormal CD276 expression is seen in several human cancers, such as HCC45. Growing evidence suggests that CD276 plays a significant role in both innate and adaptive immunity, as well as in tumor aggressiveness. It is expressed in immune cells like macrophages and APCs, regulating T cell function and contributing to cancer cell migration and invasion46. In fact, data obtained in vitro and in vivo by Liu et al. have demonstrated that, in HCC patients, high levels of CD276 are associated with poor prognosis, modifications in immune cell infiltration, immune marker expression, and macrophage polarization5. Authors concluded that CD276 could be a possible prognostic marker and play a role as a putative target for immunotherapy in HCC. Furthermore, Cheng et al. have described a new pivotal role of CD276 in HCC. Based on their data, CD276 promotes vasculogenic mimicry formation in HCC via the PI3K/AKT/MMPs pathway44. Vasculogenic mimicry is a process put in action by tumor to obtain sufficient blood supply without involving endothelial cell. This process is one of the hallmarks of cancer and it is closely linked to invasion, metastasis and poor outcome47. Interestingly, CD276 can activate matrix metalloproteinases (MMPs), in particular MMP14/MMP2 and MMP2/MMP9, suggesting that it can both regulate tumor aggressiveness and the EMT process and improve vasculogenic mimicry formation44.
Unfortunately, CD276 can be detected only by mRNA assay, and this nullifies its use as a serum marker. However, its potential as a therapeutic target remains of interest, since interfering with the EMT process and tumor invasion could have significant antitumor effects.
FGF19
Fibroblast growth factor (FGF) 19 is a member of FGFs family. When these proteins link their receptor, the FGF receptor (FGFR) tyrosine kinase can regulate multiple biological processes such as cell growth, differentiation, angiogenesis, and metabolism48,49,50,51. The principal receptor for FGF19 is FGFR4, that is able to form a selective linking52. In healthy conditions, FGF19 regulates bile acid synthesis and nutrient metabolism48. Several studies have demonstrated that the FGF19-FGFR4 axis plays a role to promote tumorigenesis, and high expression of FGF19 indicates a poor prognosis in several cancers, including primary liver cancers48,49,50,51. In 2012, Miura et al. suggested HCC expression of FGF19 mRNA as an independent prognostic factor for overall survival and disease-free survival53. Using the Cancer Genome Atlas (TCGA) database, around 7% of HCC patients were found to have a high copy number of FGF19, and these patients showed shorter median survival compared with the low-expression cohort54. To evaluate Fisogatinib (BLU-554) in a phase I study, Kim et al. have divided their patients on the basis of FGF19 expression measured by IHC assay. Interestingly, Authors demonstrated that FGF19 is expressed in HCC tissues, but it was not detected in adjacent normal liver49. Consistently, Maeda et al. reported that serum FGF19 levels are significantly elevated in HCC patients compared to healthy subjects and patients with chronic liver disease. However, serum FGF19 levels alone showed diagnostic performance comparable to that of AFP or DCP levels. Remarkably, when the authors analyzed these three serum biomarkers together, they achieved high sensitivity, suggesting that this combination could aid in the diagnosis of HCC, particularly in cases of small tumors50.
These findings highlight the potential of FGF19 as a biomarker for HCC diagnosis, especially when used in combination with other biomarkers like AFP and DCP. However, additional studies should be performed to validate its clinical use.
INMT
Indolethylamine N-methyltransferase (INMT), also known as amine N-methyltransferase, is an enzyme that catalyzes the methylation of thioether and selenoether compounds using S-adenosylmethionine (SAM) as a methyl donor55,56. Recently, it has been proposed as a prognostic marker due to the progressive decrease in its mRNA and protein levels across liver cancer stages. In fact, two independent works studying two different pathways have detected a strong INMT mRNA reduction in both murine and human HCC samples, confirmed by using data deposited in The Cancer Genome Atlas (TCGA) database55,56. Torres et al. studied DNA damage induced by diethylnitrosamine in the liver, promoting carcinogenic effects mediated by Nuclear Factor Erythroid 2–Like 2 (NRF2). During their analysis, the authors found that INMT was significantly downregulated in both animal models and human tissues. Moreover, INMT mRNA levels, obtained from the TCGA database, were closely associated with poor overall survival in patients55. Similarly, Sun et al. mRNA analysis indicated that low INMT levels correlate with poor prognosis and overall survival of HCC patients56. In their studies, Authors also confirmed that INMT expression is low in liver cancer, and, in in vitro experiments, they demonstrated that colony formation and cell proliferation were strongly increased by knockdown of INMT56. Sun et al. have also studied the role of INMT in hepatic selenium metabolism. Selenium is a fundamental mineral for human health, and the liver is the principal organ that suffers from selenium deficiency, being selenium levels negatively associated with liver diseases57.
Interestingly, the same enzyme, analyzed in two different metabolic contexts, shows consistent results in HCC patients, suggesting that INMT could serve as a prognostic marker for HCC. However, the underlying molecular pathways remain unknown.
MMP10
Matrix metallopreonases (MMPs) are proteases capable of degrading all kinds of extracellular matrix proteins. Their role as tumoral microenvironment modulators is widely documented58. MMP10, strongly upregulated in liver injury conditions, is able to degrade several components of the extracellular matrix and it is capable to activate other MMPs59. Recently, this molecule has been correlated with HCC prognosis and overall survival by gene analysis of several human cohorts60,61. García-Irigoyen et al. firstly demonstrated that MMP10 plays a role in HCC development, tumor angiogenesis and tumor growth. Furthermore, Authors demonstrated that MMP10 is involved in the C-X-C chemokine receptor-4 (CXCR4) / stromal-derived factor-1 (SDF1) axis, enhancing metastasis and progression processes by the mitogen-activated protein kinase/extracellular signal-regulated kinase (ERK) 1/2 (MEK-ERK1/2) pathway59. In this same study, increased HCC MMP10 mRNA and protein expression were correlated with poor patient prognosis59. A similar result was obtained by Liu et al.61. Using TCGA and cBioportal as Cancer Genomics databases, they identified six genes, including MMP10, as useful to predict overall survival of HCC patients61. A different approach was used by Shaglouf et al., who focused on the differential expression protein and mRNA expression in liver of Wistar rats in which they induce HCC by administration of diethyl nitrosamine (DEN) and 2-acetylaminofluorene (2-AFF) compared to control rat, and the confirmed their results by analyzing human HCC samples60. By these means, Authors found that MMP10, among other molecules, plays a pivotal regulatory role in HCC development and progression60.
Although the role of MMP10 in HCC has only been described in recent years, the literature consistently supports its major involvement in HCC progression and its potential use as a prognostic marker and predictor of overall survival. However, its assessment -whether through mRNA expression or immunohistochemistry (IHC)- requires a biopsy sample for analysis. Furthermore, as suggested by Liu et al., these findings should be validated in independent cohorts.
DKK1
Dickkopf-1 (DKK1) is a small protein, widely recognized for its role as an inhibitor of the Wnt/β-catenin pathway. Depending on the cell lines used in in vitro experiments, conflicting views exist regarding its oncogenic or tumor-suppressive function62,63,64. Interestingly, in 2011, Tung et al. proposed DKK1 as a potential serum diagnostic and prognostic biomarker for HCC patients. Moreover, DKK1 levels have been observed to increase with the severity of liver disease and the progression of HCC tumor stage55, although it did not emerge as an independent prognostic marker. However, DKK1 played a pivotal role in tumor formation and growth in a mouse animal model55. A major limitation of this study was the small sample size analyzed. The following year, in 2012, Shen et al. expanded the sample size and used AFP as a control and validation model56. Their data suggest that DKK1 could be a useful biomarker for early HCC diagnosis; however, this serum marker is not capable of distinguishing between early-stage HCC and advanced HCC. Nevertheless, the authors proposed that DKK1 could aid in the early diagnosis of patients who are AFP-negative56. Interestingly, Chen et al. investigated the role of DKK1 in HCC invasion and migration. Their findings revealed that DKK1 plays a fundamental role in promoting invasion and migration processes in HCC through the β-catenin pathway, particularly by modulating MMP7, a downstream target gene57. Supporting this evidence, Suda et al. recently demonstrated that DKK1 contributes to remodeling the HCC tumor microenvironment by influencing angiogenesis58.
The main limitation regarding the clinical use of DKK1 in the diagnosis and/or prognosis of HCC stems from its controversial biological role, which could be at least partially attributed to the different types of cell lines used in experiments in available studies. Nonetheless, in recent years, many researchers have focused on identifying the pathways and specific molecules involved, aiming to better understand the potential clinical applications of DKK1.
OPN
Osteopontin (OPN) is a glycoprotein involved in various physiological and pathological processes, including inflammation, immunity, and tumor progression65. Using a small Egyptian cohort, Abdel-Hafiz et al. demonstrated that OPN is overexpressed in HCC serum samples compared to healthy subjects. Moreover, authors observed a correlation between OPN levels analyzed by IHC and tumor grade66. Interestingly, OPN levels were negatively correlated with liver function, making the authors suggest OPN as a prognostic biomarker66. In the same period, Zhu et al. used a bigger Asian cohort and showed that OPN could be a good biomarker for HCC monitoring. In fact, authors detected serum OPN in HCC patients with tumor smaller than 2 cm, showing a higher sensitivity with respect to AFP67. The limitation of both cohorts was that most patients had HBV-related liver disease.
Several research teams have demonstrated the prognostic value of OPN in HCC. High OPN expression has been linked to poor prognosis, as it is involved in promoting angiogenesis, tumor cell migration, and invasion—key processes in cancer metastasis68,69. Wang et al. analyzed HCC tissues by HCC obtained from the surgery resection of patients without chemo- and/or radio-therapy treatment. By their data, OPN levels in HCC patients correlated with increase vascular invasion, tumor stages, reduced overall survival and disease-free survival70. As a confirmation of these data, Zhu et al. proved that mice KO for OPN are more resistant to HCC development than WT mice when exposed to DEN and carbon tetrachloride71. In their work, authors demonstrated the connection between macrophage migration, programmed death ligand 1 (PD-L1) expression, and OPN in a mouse animal model. This occurs through activation of the colony-stimulating factor-1 (CSF1) – CSF1 receptor (CSF1R) pathway in macrophages71.
All available human data support the potential use of OPN as a clinical serum biomarker for the early diagnosis and prognosis of HCC. However, the limited cohort sizes and the predominance of HBV-associated HCC cases highlight the need for further studies before recommending OPN for clinical application. Concurrently, clinical observations and correlations with OPN levels in HCC tissues underscore its crucial role in HCC malignancy, tumor formation, and metastasis.
RNA sequencing
Over the past decade, advancements in single-cell sequencing technologies have revolutionized our understanding of cancer biology, particularly in the context of HCC. HCC is characterized by significant intratumoural heterogeneity, which complicates diagnosis, prognosis, and treatment. Recent studies utilizing single-cell RNA sequencing (scRNA-seq) have revealed distinct subpopulations of cells within HCC tumors, including various types of cancer cells, immune cells, and stromal cells. Based on this idea, Zhang et al. performed a landmark study, using single-cell sequencing to classify HCC tumors into distinct subtypes based on their immune microenvironment72. Moreover, Zheng et al. identified a unique subset of HCC cancer stem cells (CSCs) that exhibit high levels of stemness and are associated with poor prognosis. These CSCs are thought to drive tumor initiation, progression, and resistance to therapy, making them crucial targets for prognostic assessment and therapeutic intervention73. Ma et al. demonstrated that scRNA-seq could help identifying distinct gene expression signatures in HCC that are not detectable by conventional methods. These signatures include markers associated with tumor aggressiveness, metastatic potential, and immune evasion. Authors found that vascular endothelial growth factor (VEGF) could play a key role in driving HCC progression74. Several researchers have focused their work on the immune landscape of HCC as a prognostic model. Indeed, the analysis of immune populations in HCC reveals significant heterogeneity of immune cells within the tumor microenvironment, encompassing diverse subsets of T cells, macrophages, and natural killer cells, each contributing uniquely to tumor progression or suppression73,74.
However, this technology is not without significant limitations. The enzymatic or mechanical tissue dissociation required for scRNA-seq can introduce transcriptional artifacts and lead to a biased representation of the tissue, often causing the underrepresentation of fragile cell types such as mature hepatocytes. A major drawback is the complete loss of spatial information, which prevents the analysis of how different cell types are organized and interact within the tumor microenvironment (TME)75,76. Furthermore, standard library preparation protocols based on poly-A selection exclude non-polyadenylated transcripts (e.g., certain non-coding RNAs) that may have crucial regulatory roles in cancer progression. To overcome some of these challenges, newer methodologies have emerged. Single-nucleus RNA sequencing (snRNA-seq) bypasses the harsh dissociation step by profiling RNA from nuclei isolated from fresh or frozen tissue. This preserves a more accurate representation of cellular diversity and allows for the capture of cell types and states that are often lost during scRNA-seq protocols77. In parallel, spatial transcriptomics technologies are directly addressing the need for spatial context. While often providing less transcriptional depth compared to single-cell methods, they map gene expression directly onto the tissue architecture. This allows for the study of cellular “neighborhoods” and the identification of spatially defined functional niches within the TME. Advanced imaging-based spatial methods, such as Multiplexed Error-Robust Fluorescence In Situ Hybridization (MERFISH), provide highly multiplexed, spatially resolved single-cell analysis at subcellular resolution78. The true frontier now lies in the multimodal integration of these technologies. By computationally combining the deep molecular profiling of scRNA-seq or snRNA-seq with the spatial maps from transcriptomics, it is possible to achieve a unified view that links cellular identity and state with a precise location within the tissue. Computational tools like Tangram or Cell2location enable this integration, allowing for the resolution of complex cell-cell interactions, the understanding of the TME’s functional logic, and the discovery of regional biomarkers76,79. This integrated approach is essential for advancing diagnostic and therapeutic strategies in HCC.
Liquid biopsy
Liquid biopsy refers to the analysis of tumor-derived components, such as circulating tumor DNA (ctDNA), circulating tumor cells (CTCs), microRNAs (miRNAs), and exosomes, present in the blood or other body fluids80. ctDNA is fragmented DNA released into the bloodstream by apoptotic or necrotic tumor cells. It carries tumor-specific genetic and epigenetic alterations, making it a valuable biomarker for cancer diagnosis and/or prognosis80. Studies have demonstrated that ctDNA can detect genetic mutations commonly associated with HCC, such as those in the TERT promoter, TP53, and CTNNB1 genes80,81,82. Additionally, hypermethylation of the RASSF1A and SEPT9 genes in ctDNA has shown promise in distinguishing HCC patients from those with benign liver diseases or healthy controls83,84. CTCs are rare cancer cells that have shed from the primary tumor into the bloodstream80. In HCC, the presence of CTCs has been associated with advanced disease stages and poor prognosis. The molecular profiling of CTCs can reveal key mutations, especially those expressing epithelial-mesenchymal transition (EMT) markers, and phenotypic characteristics that may aid in the early detection of HCC80,85,86,87. miRNAs are small non-coding RNAs that regulate gene expression and play critical roles in cancer development and progression80. Pellizzaro et al. have identified miR-122 highly expressed in HCC patients when compared to healthy individuals, suggesting it as a prognostic marker88. Moreover, Jin et al. found that four miRNAs (miR-1972, miR-193a-5p, miR-214-3p and miR-365a-3p) are upregulated in HCC patients and that their elevated expression could be used to distinguish them from controls89. Exosomes are small extracellular vesicles that carry proteins, lipids, and nucleic acids, including miRNAs, and play a role in cell-cell communication80. Xue et al. have demonstrated that elevated levels of exosomal miR-93 are linked to poor prognosis in HCC patients90.
Liquid biopsy represents a transformative approach in the diagnosis and prognosis of HCC, offering a non-invasive, real-time method to capture tumor dynamics. Beyond diagnosis and prognosis, liquid biopsy may also play a crucial role in guiding therapeutic decisions. ctDNA and CTC analysis can help stratify patients based on actionable genetic alterations, which may influence sensitivity or resistance to systemic therapies, including tyrosine kinase inhibitors and immune checkpoint inhibitors.
However, the sensitivity and specificity of liquid biopsy assays still require improvement to ensure reliability in clinical practice, along with further standardization and validation in large cohorts. A major challenge remains the difficulty of linking detected biomarkers to specific pathophysiological processes occurring within the liver and assigning them to distinct cellular subtypes, which limits their mechanistic interpretation80.
In this context, extracellular vehicles (EVs), including exosomes, represent a promising tool as they carry cell-specific molecular cargo (e.g., proteins, lipids, and nucleic acids) that may help trace the cellular origin of the signal, potentially bridging the gap between circulating biomarkers and intrahepatic oncogenic events80.
To fully leverage these biomarkers, it is crucial to integrate them within a broader framework of molecular and genetic classifications. By combining these novel markers with insights into HCC’s molecular subtypes, we are going to enhance our understanding of tumor heterogeneity, optimize patient stratification, and develop more targeted therapeutic approaches.
Molecular and genetic classification of HCC
Although promising roles as molecular biomarkers are described in this review, no one is still applied in clinical practices for the management of HCC. Traditional classification based on histology and staging has proven insufficient in capturing the molecular complexity of this neoplasia, which significantly influences patient prognosis and treatment outcomes91. Based on this, several teams focused on classifying HCC based on its gene and molecular expression in order to suggest different treatments for different clusters of HCC patients. Gene expression profiling has emerged as a key tool in distinguishing different molecular subtypes of HCC. Techniques such as RNA sequencing and high-throughput microarrays enable comprehensive insights into gene activation and suppression patterns across HCC tumors, distinguishing molecular characteristics associated with various oncogenic processes92. Numerous studies have proposed classification systems based on the expression of genes related to cell cycle regulation, immune response, metabolic pathways, and oncogenic signaling91,92,93. Molecular classification based on gene expression has identified several subtypes, each linked to distinct biological processes and clinical trajectories, including cell cycle dysregulation, immune response variations, and metabolic changes. Recent advances in genomic technologies, particularly high-throughput gene expression profiling, have revolutionized HCC classification, leading to the identification of distinct molecular subtypes that offer better insights into disease pathogenesis and therapeutic vulnerabilities91,92,93. While molecular classification allows for nuanced understanding and targeted intervention, implementing these insights into clinical practice remains challenging. High costs, the need for complex bioinformatics support, and the rapid evolution of molecular profiling technology present obstacles in making these classifications universally available. Nonetheless, these classifications have shown potential in guiding more personalized treatment, aiding in early-stage patient stratification, and enabling closer monitoring for relapse risks93. For instance, the inflammation subtype and the immune class subtype of HCC, characterized by high expression of immune checkpoint molecules, are more likely to respond to immune checkpoint inhibitors. Conversely, tumors with Wnt/β-catenin activation exhibit an immunosuppressive microenvironment and may be resistant to ICIs. Therefore, molecular classification can serve as a valuable tool for patient stratification, enabling a more personalized therapeutic approach.
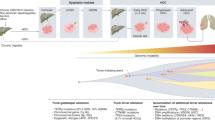
Below, we will report several prominent molecular classification models, focusing on those proposed in recent high-impact studies, a summary of which is presented in Fig. 3.
A Overview of the principal molecular and genetic classification systems proposed for HCC, with emphasis on their defining criteria and conceptual focus. B Timeline outlining the historical emergence and origin of each classification framework included in this study. Created with BioRender.com (https://BioRender.com/fdrtpgo).
The proliferation and inflammation subtypes
One of the earliest classification models for HCC divides it into Proliferation and Inflammation subtypes, based on the differential expression of genes associated with cell proliferation and immune response94,95. The Proliferation subtype is characterized by the overexpression of genes involved in cell cycle regulation and oncogenic pathways, particularly Wnt/β-catenin and MYC signaling. This subtype is associated with a poor prognosis, including aggressive tumor growth and a high likelihood of vascular invasion93. The Inflammation subtype, by contrast, displays overexpression of immune-related genes, including those involved in interferon signaling pathways, which correlate with a relatively favorable prognosis. This subtype’s immune-enriched microenvironment makes it potentially responsive to immune-modulating therapies, particularly checkpoint inhibitors targeting PD-1 and CTLA-496.
For the Proliferation subtype, targeted therapies aimed at inhibiting the Wnt/β-catenin and MYC pathways are under investigation. Small molecule inhibitors targeting MYC and Wnt/β-catenin are being explored, though challenges in drug development for these targets still remain97. The potential of the Inflammation subtype for immune checkpoint therapy has spurred clinical trials investigating PD-1 inhibitors, such as pembrolizumab, which has demonstrated promising results in HCC patients with immune-enriched tumor environments98.
The Cancer Genome Atlas (TCGA) Classification: S1-S4 Subtypes
The Cancer Genome Atlas (TCGA) provided a comprehensive framework for classifying HCC into four molecular subtypes, known as S1-S4. This classification system offers deeper insights into the underlying genomic and epigenomic landscape of HCC tumors99. Each subtype is characterized by distinct gene expression profiles, reflecting different molecular pathways and clinical behaviors:
-
S1 subtype: This subtype is characterized by activation of the Wnt/β-catenin pathway and metabolic dysregulation, often with a favorable prognosis due to its association with less aggressive clinical features.
-
S2 subtype: Marked by TP53 mutations and chromosomal instability, S2 is associated with high genomic instability and poor outcomes. TP53 mutations in this subtype often lead to resistance to certain chemotherapies, necessitating alternative approaches.
-
S3 subtype: Characterized by Ras/MAPK signaling activation, this subtype exhibits frequent growth factor receptor mutations, which can be targeted by tyrosine kinase inhibitors.
-
S4 subtype: Featuring an immune-rich microenvironment, S4 shows high expression of immune checkpoint molecules such as PD-1 and CTLA-4, and is potentially responsive to immunotherapy, offering a more favorable outlook for patients with advanced HCC.
The S4 subtype’s immune signature makes it an ideal candidate for checkpoint inhibitors, which have shown significant promise in recent clinical trials. Conversely, S2 and S3 subtypes may benefit from therapies that address chromosomal instability and receptor-mediated pathways, such as tyrosine kinase inhibitors targeting growth factor receptors in S399.
iCluster subtypes: integrative molecular subtypes
Wheeler and Roberts introduced an integrative molecular approach through iCluster analysis, which combines gene expression data with epigenetic and proteomic information. This integrative classification is especially beneficial in cases where single-layer analysis (e.g., genomics alone) may not capture the full spectrum of tumor biology99. This approach revealed three main subtypes of HCC:
-
iCluster 1: Enriched for chromatin remodeling genes and metabolic dysregulation, with a poor prognosis. This cluster showed a low frequency of cyclin-dependent kinase inhibitor 2 A (CDKN2A) silencing, mutation in CTNNB1 and TERT promoter, a reduced TERT expression, and, simultaneously, a high proliferation marker genes expression.
-
iCluster 2: Marked by immune activation and stromal features, this group has a relatively favorable prognosis and may benefit from immunotherapy. It was characterized by lower-grade tumors with less microvascular invasion, suggest that these tumors may respond well to immunotherapy.
-
iCluster 3: This group shows high activation of oncogenic signaling pathways, including Wnt/β-catenin and TGF-β pathways, chromosome instability, multiple CpG sites, and is associated with aggressive tumor characteristics and poor survival.
iCluster 1’s chromatin remodeling features may be targeted with drugs designed to modulate epigenetic pathways. Meanwhile, iCluster 2 could benefit from immune-based therapies due to its immune-rich characteristics. iCluster 3, with its reliance on oncogenic signaling, may respond to TGF-β inhibitors or combination regimens targeting multiple pathways.
CIMP and non-CIMP subtypes
Li et al. categorized HCC based on the CpG island methylator phenotype (CIMP), a distinct epigenetic signature associated with hypermethylation of CpG islands in gene promoter regions100. The CIMP subtype is characterized by extensive DNA methylation and poor prognosis. This epigenetic subtype (CIMP subtype) is gaining attention as a potential biomarker for identifying patients who may benefit from novel epigenetic therapies targeting DNA methylation and chromatin remodeling enzymes.
The hypermethylation patterns in CIMP make this subtype a candidate for drugs like DNA methyltransferase inhibitors. Ongoing trials are exploring these drugs for HCC, providing a potential novel therapeutic option for patients with aggressive epigenetic profiles.
Immune subtypes
In addition to the Inflammation subtype described by Llovet et al., another classification by Sia and collaborators focused on immune-specific subtypes96. Using the potential of omics technologies, in particular the sequencing of the whole RNA of a tissue, they divided HCC patients according to different TME characteristics and they identified a novel “Immune class,” comprising approximately 25% of cases, which exhibits heightened immune cell infiltration, elevated expression of immune regulatory molecules, and markers indicative of cytotoxic T-cell activity. This Immune class is further divided into two distinct subclasses based on immune microenvironment characteristics: an Active Immune Response subtype, associated with a favorable prognosis and enriched in adaptive immune signaling, and an Exhausted Immune Response subtype, marked by poor prognosis due to immunosuppressive signaling and T-cell exhaustion96.
The Exhausted Immune subtype demonstrated upregulation of pathways associated with immune tolerance, including TGF-β signaling, which drives T-cell exhaustion and M2 macrophage polarization, potentially promoting tumor progression. Moreover, this subgroup exhibited high glycolytic activity and metabolic reprogramming, reinforcing its immunosuppressive phenotype. The findings suggest that patients within this subtype may benefit from combination therapies targeting immune checkpoints alongside TGF-β inhibitors, potentially restoring anti-tumor immunity by reversing T-cell exhaustion. In contrast, the Active Immune Response subtype showed enhanced interferon (IFN)-γ signaling and cytotoxic gene expression, suggesting a pre-existing adaptive immune response that may facilitate responsiveness to PD-1/PD-L1 blockade therapies. This subtype aligns with the immune profiles seen in other immunotherapy-responsive tumors, such as melanoma, reinforcing its potential as a target for checkpoint inhibition96.
This immune classification not only deepens understanding of HCC’s immune landscape but also offers a potential biomarker for stratifying patients in clinical settings.
Metabolic subtypes
Yang et al. described a classification based on metabolic gene expression profiles, revealing subtypes with distinct metabolic pathways101. They are divided into three subtypes (C1, C2, and C3). C1 (active) was characterized by enrichment in signatures related to amino acid, lipid, and drug metabolism, showing low AFP expression, early pathologic stages (I/II), and lower histologic grades (G1/G2), which correlated with a favorable prognosis. C2 (exhausted) lacked distinct metabolic signatures but exhibited high levels of immune and stromal cell infiltration. C1 showed a strong association with metabolic activity and favorable outcomes mirrored the features of non-proliferative HCCs. In contrast, C2 showed elevated immune infiltration, making it potentially responsive to immune checkpoint inhibitors and chemotherapy. C3, which had elevated AFP levels and a worse prognosis, showed reduced metabolic activity compared to C1 but was still more metabolically active than C2. C3 was particularly enriched in hormone and proteoglycan metabolism, indicating an intermediate state with poor prognosis.
Proteomic subtypes
In early 2024, Diao et al. focused their attention on the characterization of immune subtypes and metabolic process in early-stage HCC to refine immunotherapy strategies102. Advances in omics technologies allowed the classification of HCC into subtypes based on immune activity and metabolic reprogramming, aiding in understanding HCC’s immune microenvironment. Utilizing proteomics, they identified three distinct immune subtypes (IM1, IM2, IM3), each with unique immune infiltration and metabolic profiles.
Researchers identified three immune subtypes:
-
IM1: Characterized by low immune infiltration and a favorable prognosis.
-
IM2: Displayed intermediate immune and metabolic features.
-
IM3: Showed high immune infiltration, T-cell exhaustion, elevated glycolysis, reduced bile acid metabolism, and the poorest prognosis.
Authors emphasize the connection between metabolic reprogramming and the immune microenvironment in HCC, especially the role of increased glycolysis and bile acid dysregulation in IM3. Through this classification, the study suggests that IM3 patients may benefit most from immunotherapy102. By linking metabolic features to immune activity, the findings propose new therapeutic approaches for HCC, emphasizing the need for immune subtype-specific treatments to enhance immunotherapy outcomes.
miRNA-based subtypes
MicroRNAs (miRNAs) are short, non-coding RNA molecules that regulate gene expression post-transcriptionally. Sathipati and Ho identified distinct molecular subtypes of HCC based on miRNA expression profiles103. Using a support vector machine (SVM)-HCC model, the authors identified a 23-miRNA signature associated with both early and advanced stages of HCC. The 23-miRNA signature was prioritized based on MED scores, with miRNAs having higher MED scores contributing more significantly to prediction accuracy. Functional insights into these top-ranked miRNAs were gained through Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Gene Ontology (GO) enrichment analyses, revealing their involvement in various cancer-related and non-cancer pathways. While this miRNA signature holds promise for predicting HCC stages, the authors also examined co-expressed miRNAs to further explore potential miRNAs beyond the 23-signature set. This broader analysis could provide deeper insights into the overall impact of miRNA activity on HCC progression.
Integration with clinical staging
Emerging molecular and genetic classification frameworks offer a more detailed understanding of the heterogeneity of HCC and could complement and improve upon existing clinical staging systems, particularly Barcelona Clinic Liver Cancer (BCLC) algorithm1, which is still fundamental to guiding therapeutic decisions. While the BCLC system uses tumor burden, liver function, and patient performance status to predict prognosis and allocate treatment, molecular subtyping provides an extra biological layer that could improve risk stratification and support personalized treatment. Integrating these molecular insights into BCLC staging could enable more nuanced prognostication and better-informed treatment selection. Finally, while current clinical guidelines increasingly acknowledge the relevance of this translational approach, prospective validation studies are still needed to standardize the use of molecular classifiers in routine clinical practice.
Conclusions and future direction
HCC remains a tumor whose early diagnosis and prognosis are complex and often suboptimal using the markers currently described in guidelines. In our review, we focused on the critical need for, and recent advancements in, novel biomarkers for these purposes in patients at risk of or affected by HCC. The development of such biomarkers is essential in routine clinical practice, playing a pivotal role in improving patient outcomes and reducing healthcare costs. Recent studies have explored promising candidates like DCLK1, GPC3, CD276, FGF19, INMT, MMP10, DKK1, OPN, and innovative approaches such as single-cell sequencing and liquid biopsy. However, it is crucial to acknowledge that HCC should be understood as a complex family of tumors, exhibiting diverse histological and mutational features. This inherent heterogeneity means that some proposed markers may be suitable for one subtype but not for another. Classification based on gene and molecular expression profiling has provided valuable insights into this biological diversity, significantly improving the understanding of HCC and offering a framework for targeted therapies. Nevertheless, recent molecular and genetic subclassifications further complicate the study and identification of broadly usable markers for diagnosing, monitoring HCC development, and assessing the efficacy of implemented therapies.
In conclusion, while emerging biomarkers and technologies hold considerable promise for enhancing HCC management, continued and focused research is essential. Future directions must involve not only validating these candidates but also improving accessibility for clinical applications. The path towards a genuine advancement in HCC treatment lies in the implementation of personalized medicine. This necessitates a cohesive strategy that integrates the use of highly efficient biomarkers with thorough analyses of the specific genetic and molecular (including metabolomic) classifications unique to each patient’s HCC subtype. Such an integrated approach is paramount to tailoring therapies effectively and ultimately improving patient outcomes.
Data availability
No datasets were generated or analyzed during the current study.
References
European Association for the Study of the, L. EASL Clinical Practice Guidelines on the management of hepatocellular carcinoma. J. Hepatol. 82, 315–374 (2025).
Llovet, J. M. et al. Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 7, 6 (2021).
Lee, Y. T. et al. Risk stratification and early detection biomarkers for precision HCC screening. Hepatology 78, 319–362 (2023).
Singal, A. G. et al. AASLD Practice Guidance on prevention, diagnosis, and treatment of hepatocellular carcinoma. Hepatology 78, 1922–1965 (2023).
Liu, W. F. et al. CD276 Promotes an inhibitory tumor microenvironment in hepatocellular carcinoma and is associated with poor prognosis. J. Hepatocell. Carcinoma 11, 1357–1373 (2024).
Tavaglione, F. et al. Development and Validation of a Score for Fibrotic Nonalcoholic Steatohepatitis. Clin. Gastroenterol. Hepatol. 21, 1523–1532 e1 (2023).
Huang, D. Q., El-Serag, H. B. & Loomba, R. Global epidemiology of NAFLD-related HCC: trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 18, 223–238 (2021).
Kanwal, F. et al. Risk of hepatocellular cancer in patients with non-alcoholic fatty liver disease. Gastroenterology 155, 1828–1837 e2 (2018).
Ronot, M., Nahon, P. & Rimola, J. Screening of liver cancer with abbreviated MRI. Hepatology 78, 670–686 (2023).
Lyshchik, A. et al. Liver imaging reporting and data system contrast-enhanced US Nonradiation Treatment Response Assessment Version 2024. Radiology 311, e232369 (2024).
Singal, A. G., Ng, M. & Kulkarni, A. Advancing surveillance strategies for hepatocellular carcinoma: a new era of efficacy and precision. J. Clin. Exp. Hepatol. 14, 101448 (2024).
Pinero, F., Dirchwolf, M. & Pessoa, M. G. Biomarkers in hepatocellular carcinoma: diagnosis, prognosis and treatment response assessment. Cells 9, 1–26 (2020).
Guan, M. C. The performance of GALAD score for diagnosing hepatocellular carcinoma in patients with chronic liver diseases: a systematic review and meta-analysis. J. Clin. Med 12, 1–15 (2023).
Tayob, N. et al. Validation of the updated hepatocellular carcinoma early detection screening algorithm in a community-based cohort of patients with cirrhosis of multiple etiologies. Clin. Gastroenterol. Hepatol. 19, 1443–1450.e6 (2021).
El-Serag, H. B. et al. HES V2.0 outperforms GALAD for detection of HCC: A phase 3 biomarker study in the United States. Hepatology 81, 465–475 (2025) .
He, H. et al. A stratified precision screening strategy for enhancing hepatitis B- and C-associated liver cancer detection: a prospective study. Sci. Rep. 15, 11396 (2025).
Miick, R., Minimo, C. & Bombonati, A. Hepatocellular Carcinoma Pathology, in Hepato-Pancreato-Biliary Malignancies: Diagnosis and Treatment in the 21st Century, C. Doria and J.N. Rogart, Editors. 2022, Springer International Publishing: Cham. p. 49–86.
Torbenson, M. S. Hepatocellular carcinoma: making sense of morphological heterogeneity, growth patterns, and subtypes. Hum. Pathol. 112, 86–101 (2021).
Koch, P. S. et al. Angiodiversity and organotypic functions of sinusoidal endothelial cells. Angiogenesis 24, 289–310 (2021).
Choi, J. H. & Thung, S. N. Advances in histological and molecular classification of hepatocellular carcinoma. Biomedicines, 11, (2023).
Nevi, L. et al. DCLK1, a putative stem cell marker in human Cholangiocarcinoma. Hepatology 73, 144–159 (2021).
Velazquez-Enriquez, J. M. et al. DCLK1 is Overexpressed and associated with immune cell infiltration in hepatocellular carcinoma. Biochem Genet. 4280–4302 (2024).
Ali, N. et al. Inflammatory and oncogenic roles of a tumor stem cell marker doublecortin-like kinase (DCLK1) in virus-induced chronic liver diseases. Oncotarget 6, 20327–20344 (2015).
Chandrakesan, P. et al. Dclk1, a tumor stem cell marker, regulates pro-survival signaling and self-renewal of intestinal tumor cells. Mol. Cancer 16, 30 (2017).
Li, J. et al. Doublecortin-Like Kinase 1 (DCLK1) Regulates B Cell-Specific Moloney Murine Leukemia Virus Insertion Site 1 (Bmi-1) and is associated with metastasis and prognosis in pancreatic cancer. Cell Physiol. Biochem 51, 262–277 (2018).
Lv, Y. et al. Doublecortin-like kinase 1 is a novel biomarker for prognosis and regulates growth and metastasis in basal-like breast cancer. Biomed. Pharmacother. 88, 1198–1205 (2017).
Panneerselvam, J. et al. DCLK1 Regulates tumor stemness and cisplatin resistance in non-small cell lung cancer via ABCD-Member-4. Mol. Ther. Oncol. 18, 24–36 (2020).
Westphalen, C. B. et al. Dclk1 Defines quiescent pancreatic progenitors that promote injury-induced regeneration and tumorigenesis. Cell Stem Cell 18, 441–455 (2016).
Weygant, N. et al. DCLK1 is a broadly dysregulated target against epithelial-mesenchymal transition, focal adhesion, and stemness in clear cell renal carcinoma. Oncotarget 6, 2193–2205 (2015).
Broner, E. C. et al. Doublecortin-Like Kinase 1 (DCLK1) Is a Novel NOTCH pathway signaling regulator in head and neck squamous cell carcinoma. Front Oncol. 11, 677051 (2021).
Chhetri, D. et al. Pleiotropic effects of DCLK1 in cancer and cancer stem cells. Front Mol. Biosci. 9, 965730 (2022).
Qu, D. et al. Overexpression of DCLK1-AL increases tumor cell invasion, drug resistance, and KRAS activation and can be targeted to inhibit tumorigenesis in pancreatic cancer. J. Oncol. 2019, 6402925 (2019).
Wu, X. Cancer Stem Cell Marker DCLK1 Correlates with Tumorigenic Immune Infiltrates in the Colon and Gastric Adenocarcinoma Microenvironments. Cancers 12, 1–15 (2020).
Harada, Y. et al. Prognostic impact of doublecortin-like kinase 1 expression in locally advanced rectal cancer treated with preoperative chemoradiotherapy. APMIS 126, 486–493 (2018).
Gao, T. et al. DCLK1 is up-regulated and associated with metastasis and prognosis in colorectal cancer. J. Cancer Res Clin. Oncol. 142, 2131–2140 (2016).
Makino, S. et al. DCLK1 integrates induction of TRIB3, EMT, drug resistance and poor prognosis in colorectal cancer. Carcinogenesis 41, 303–312 (2020).
Moore, L. L. From inflammation to oncogenesis: tracing serum DCLK1 and miRNA Signatures in chronic liver diseases. Int. J. Mol. Sci 25, 1–20 (2024).
Yan, R. et al. DCLK1 suppresses tumor-specific Cytotoxic T Lymphocyte function through recruitment of MDSCs via the CXCL1-CXCR2 Axis. Cell Mol. Gastroenterol. Hepatol. 15, 463–485 (2023).
Zhao, Q. et al. Crosstalk between colorectal CSCs and immune cells in tumorigenesis, and strategies for targeting colorectal CSCs. Exp. Hematol. Oncol. 13, 6 (2024).
An, S. et al. GPC3-targeted immunoPET imaging of hepatocellular carcinomas. Eur. J. Nucl. Med Mol. Imaging 49, 2682–2692 (2022).
Li, S. Q. et al. Prediction of glypican-3 expression in hepatocellular carcinoma using multisequence magnetic resonance imaging-based histology nomograms. Quant. Imaging Med Surg. 14, 4436–4449 (2024).
Cao, B. et al. Development of GPC3-CAR-NK cells and optimization as a therapy for HCC. J Leukoc Biol 117, qiae144 (2024).
Li, H. et al. Preclinical and clinical studies of CAR-NK-cell therapies for malignancies. Front Immunol. 13, 992232 (2022).
Cheng, R. et al. CD276 Promotes Vasculogenic Mimicry Formation in Hepatocellular Carcinoma via the PI3K/AKT/MMPs Pathway. Onco Targets Ther. 13, 11485–11498 (2020).
Kontos, F. et al. B7-H3: An attractive target for antibody-based immunotherapy. Clin. Cancer Res 27, 1227–1235 (2021).
Liu, S. et al. The Role of CD276 in cancers. Front Oncol. 11, 654684 (2021).
Luo, Q. et al. Vasculogenic mimicry in carcinogenesis and clinical applications. J. Hematol. Oncol. 13, 19 (2020).
Uriarte, I. et al. Ileal FGF15 contributes to fibrosis-associated hepatocellular carcinoma development. Int J. Cancer 136, 2469–2475 (2015).
Kim, R. D. et al. First-in-Human Phase I Study of Fisogatinib (BLU-554) Validates Aberrant FGF19 Signaling as a Driver Event in Hepatocellular Carcinoma. Cancer Discov. 9, 1696–1707 (2019).
Maeda, T. et al. Serum fibroblast growth factor 19 serves as a potential novel biomarker for hepatocellular carcinoma. BMC Cancer 19, 1088 (2019).
Guo, C. et al. FGF19/FGFR4 signaling contributes to hepatocellular carcinoma survival and immune escape by regulating IGF2BP1-mediated expression of PD-L1. Biomed. Pharmacother. 170, 115955 (2024).
Raja, A. et al. FGF19-FGFR4 signaling in hepatocellular carcinoma. Cells 8, 1–14 (2019).
Miura, S. et al. Fibroblast growth factor 19 expression correlates with tumor progression and poorer prognosis of hepatocellular carcinoma. BMC Cancer 12, 56 (2012).
Chen, Z. et al. The Role of Fibroblast Growth Factor 19 in Hepatocellular Carcinoma. Am. J. Pathol. 191, 1180–1192 (2021).
Lopez-Torres, C. D. et al. Downregulation of indolethylamine N-methyltransferase is an early event in the rat hepatocarcinogenesis and is associated with poor prognosis in hepatocellular carcinoma patients. J. Gene Med 24, e3439 (2022).
Sun, H. et al. Development and validation of a selenium metabolism regulators associated prognostic model for hepatocellular carcinoma. BMC Cancer 23, 451 (2023).
Lin, Y. et al. Selenium status in patients with chronic liver disease: a systematic review and meta-analysis. Nutrients 14, 1–13 (2022).
Bassiouni, W., Ali, M. A. M. & Schulz, R. Multifunctional intracellular matrix metalloproteinases: implications in disease. FEBS J. 288, 7162–7182 (2021).
Garcia-Irigoyen, O. et al. Matrix metalloproteinase 10 contributes to hepatocarcinogenesis in a novel crosstalk with the stromal derived factor 1/C-X-C chemokine receptor 4 axis. Hepatology 62, 166–178 (2015).
Shaglouf, L. H. F. et al. Elevated expression of ISY1, APOA-1, SYNE1, MTG1, and MMP10 at HCC initiation: HCC specific protein network involving interactions of key regulators of lipid metabolism, EGFR signaling, MAPK, and splicing pathways. Protoplasma 260, 651–662 (2023).
Liu, G. M. et al. Identification of a six-gene signature predicting overall survival for hepatocellular carcinoma. Cancer Cell Int 19, 138 (2019).
Tung, E. K. et al. Clinicopathological and prognostic significance of serum and tissue Dickkopf-1 levels in human hepatocellular carcinoma. Liver Int 31, 1494–1504 (2011).
Shen, Q. et al. Serum DKK1 as a protein biomarker for the diagnosis of hepatocellular carcinoma: a large-scale, multicentre study. Lancet Oncol. 13, 817–826 (2012).
Chen, L. et al. DKK1 promotes hepatocellular carcinoma cell migration and invasion through beta-catenin/MMP7 signaling pathway. Mol. Cancer 12, 157 (2013).
Bruha, R., Vitek, L. & Smid, V. Osteopontin - A potential biomarker of advanced liver disease. Ann. Hepatol. 19, 344–352 (2020).
Abdel-Hafiz, S. M. et al. Evaluation of Osteopontin as a Biomarker in Hepatocellular Carcinomas in Egyptian Patients with Chronic HCV Cirrhosis. Asian Pac. J. Cancer Prev. 19, 1021–1027 (2018).
Zhu, M. et al. OPN is a promising serological biomarker for hepatocellular carcinoma diagnosis. J. Med Virol. 92, 3596–3603 (2020).
Panda, V. K. et al. Osteopontin: A key multifaceted regulator in tumor progression and immunomodulation. Biomedicines 12, 1–26 (2024).
Moorman, H. R. et al. Osteopontin: a key regulator of tumor progression and immunomodulation. Cancers 12, 1–18 (2020).
Wang, H. et al. Increased expression of osteopontin indicates poor prognosis in hepatocellular carcinoma. Int J. Clin. Exp. Pathol. 11, 5916–5922 (2018).
Zhu, Y. et al. Disruption of tumour-associated macrophage trafficking by the osteopontin-induced colony-stimulating factor-1 signalling sensitises hepatocellular carcinoma to anti-PD-L1 blockade. Gut 68, 1653–1666 (2019).
Zhang, Q. et al. Integrated multiomic analysis reveals comprehensive tumour heterogeneity and novel immunophenotypic classification in hepatocellular carcinomas. Gut 68, 2019–2031 (2019).
Zheng, H. et al. Single-cell analysis reveals cancer stem cell heterogeneity in hepatocellular carcinoma. Hepatology 68, 127–140 (2018).
Ma, L. et al. Tumor Cell Biodiversity Drives Microenvironmental Reprogramming in Liver Cancer. Cancer Cell 36, 418–430.e6 (2019).
Saviano, A., Henderson, N. C. & Baumert, T. F. Single-cell genomics and spatial transcriptomics: Discovery of novel cell states and cellular interactions in liver physiology and disease biology. J. Hepatol. 73, 1219–1230 (2020).
Andersson, A. et al. Single-cell and spatial transcriptomics enables probabilistic inference of cell type topography. Commun. Biol. 3, 565 (2020).
Andrews, T. S. et al. Single-Cell, Single-Nucleus, and Spatial RNA Sequencing of the Human Liver Identifies Cholangiocyte and Mesenchymal Heterogeneity. Hepatol. Commun. 6, 821–840 (2022).
Watson, B. R. et al. Spatial transcriptomics of healthy and fibrotic human liver at single-cell resolution. Nat. Commun 16, 319 (2025).
Lotfollahi, M. et al. Mapping single-cell data to reference atlases by transfer learning. Nat. Biotechnol. 40, 121–130 (2022).
Lehrich, B. M. et al. Battle of the biopsies: Role of tissue and liquid biopsy in hepatocellular carcinoma. J. Hepatol. 80, 515–530 (2024).
Kancherla, V. et al. Genomic Analysis Revealed New Oncogenic Signatures in TP53-Mutant Hepatocellular Carcinoma. Front Genet 9, 2 (2018).
Nault, J. C. et al. Clinical Impact of Genomic Diversity From Early to Advanced Hepatocellular Carcinoma. Hepatology 71, 164–182 (2020).
Li, J. et al. RASSF1A methylation as a biomarker for detection of colorectal cancer and hepatocellular carcinoma. World J. Gastrointest. Oncol. 14, 1574–1584 (2022).
Li, B. et al. SEPT9 gene methylation as a noninvasive marker for hepatocellular carcinoma. Dis. Markers 2020, 6289063 (2020).
Prasoppokakorn, T. et al. Circulating tumor cells as a prognostic biomarker in patients with hepatocellular carcinoma. Sci. Rep. 12, 18686 (2022).
Orrapin, S. et al. Clinical Implication of Circulating Tumor Cells Expressing Epithelial Mesenchymal Transition (EMT) and Cancer Stem Cell (CSC) Markers and Their Perspective in HCC: A Systematic Review. Cancers 14, 1–21 (2022).
Yu, J. J. et al. The Presence of Circulating Tumor Cell Cluster Characterizes an Aggressive Hepatocellular Carcinoma Subtype. Front Oncol. 11, 734564 (2021).
Pelizzaro, F. et al. Circulating MicroRNA-21 and MicroRNA-122 as Prognostic Biomarkers in Hepatocellular Carcinoma Patients Treated with Transarterial Chemoembolization. Biomedicines 9, 1–12 (2021).
Jin, Y. et al. Circulating microRNAs as potential diagnostic and prognostic biomarkers in hepatocellular carcinoma. Sci. Rep. 9, 10464 (2019).
Xue, X. et al. Exosomal miR-93 promotes proliferation and invasion in hepatocellular carcinoma by directly inhibiting TIMP2/TP53INP1/CDKN1A. Biochem Biophys. Res Commun. 502, 515–521 (2018).
Chidambaranathan-Reghupaty, S., Fisher, P. B. & Sarkar, D. Hepatocellular carcinoma (HCC): Epidemiology, etiology and molecular classification. Adv. Cancer Res. 149, 1–61 (2021).
Wu, Y., Liu, Z. & Xu, X. Molecular subtyping of hepatocellular carcinoma: A step toward precision medicine. Cancer Commun. (Lond.) 40, 681–693 (2020).
Rebouissou, S. & Nault, J. C. Advances in molecular classification and precision oncology in hepatocellular carcinoma. J. Hepatol. 72, 215–229 (2020).
Llovet, J. M. et al. Plasma biomarkers as predictors of outcome in patients with advanced hepatocellular carcinoma. Clin. Cancer Res 18, 2290–2300 (2012).
Lee, S.H. et al., Molecular Subtypes and Genomic Signatures of Hepatocellular Carcinoma for Prognostication and Therapeutic Decision-Making, in Hepatocellular Carcinoma: Translational Precision Medicine Approaches, Y. Hoshida, Editor. 2019: Cham (CH). p. 109–23.
Sia, D. et al. Identification of an Immune-specific Class of Hepatocellular Carcinoma, Based on Molecular Features. Gastroenterology 153, 812–826 (2017).
Liu, J. et al. Wnt/beta-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal Transduct. Target Ther. 7, 3 (2022).
Qin, S. et al. Pembrolizumab Versus Placebo as second-line therapy in patients from Asia With advanced hepatocellular carcinoma: a randomized, double-blind, Phase III trial. J. Clin. Oncol. 41, 1434–1443 (2023).
Cancer Genome Atlas Research Network Electronic address, w.b.e. and N. Cancer Genome Atlas Research, comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 169, 1327–1341.e23 (2017).
Li, G. et al. Development and validation of a CIMP-associated prognostic model for hepatocellular carcinoma. EBioMedicine 47, 128–141 (2019).
Yang, C. et al. Metabolism-associated molecular classification of hepatocellular carcinoma. Mol. Oncol. 14, 896–913 (2020).
Diao, L. et al. Identification of Proteome-based immune subtypes of early hepatocellular carcinoma and analysis of potential metabolic drivers. Mol. Cell Proteom. 23, 100686 (2024).
Yerukala Sathipati, S. & Ho, S. Y. Novel miRNA signature for predicting the stage of hepatocellular carcinoma. Sci. Rep. 10, 14452 (2020).
Krings, G. et al. Immunohistochemical pitfalls and the importance of glypican 3 and arginase in the diagnosis of scirrhous hepatocellular carcinoma. Mod. Pathol. 26, 782–791 (2013).
Calderaro, J. et al. Histological subtypes of hepatocellular carcinoma are related to gene mutations and molecular tumour classification. J. Hepatol. 67, 727–738 (2017).
Kim, H., Jang, M. & Park, Y. N. Histopathological Variants of Hepatocellular Carcinomas: an Update According to the 5th Edition of the WHO Classification of Digestive System Tumors. J. Liver Cancer 20, 17–24 (2020).
Ganeshan, D. et al. Imaging features of fibrolamellar hepatocellular carcinoma. AJR Am. J. Roentgenol 202, 544–552 (2014).
Griffith, O. L. et al. A genomic case study of mixed fibrolamellar hepatocellular carcinoma. Ann. Oncol. 27, 1148–1154 (2016).
Vij, M. et al. Clinicopathological Characteristics of Neutrophil-Rich Hepatocellular Carcinoma: An Uncommon Subtype of Primary Liver Cancer. Int. J. Surg. Pathol. 33, 828–841 (2025).
Wang, P. Y. et al. Lymphocyte-Rich Hepatocellular Carcinoma with Multiple Lymphadenopathy and Positive Epstein-Barr Virus Encoding Region. Case Reports Hepatol. 2023, 4797233 (2023).
Acknowledgements
Not applicable.
Author information
Authors and Affiliations
Contributions
Conceptualization L.N., A.B., S.C.; writing—original draft preparation L.N., A.B., S.C.; writing—review and editing L.N., C.A., A.A., P.G., U.V-G., S.C.; supervision M.P.G., U.V-G., A.B., S.C.; visualization C.A., F.M., T.M., C.T.; A.B. and S.C. share the same authorship. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Nevi, L., Aiello, C., Molinaro, F. et al. Decoding the molecular and genomic landscape of hepatocellular carcinoma: biomarker discovery, classification frameworks, and therapeutic targeting. npj Gut Liver 2, 25 (2025). https://doi.org/10.1038/s44355-025-00038-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s44355-025-00038-8
This article is cited by
-
Next-generation dynamic and combinatorial nanotherapies for liver cancer: mechanisms, current advances and future perspectives
Journal of Nanobiotechnology (2026)