Abstract
Fungal pathogenesis requires a number of extracellularly released virulence factors. Recent studies demonstrating that most fungal extracellular molecules lack secretory tags suggest that unconventional secretion mechanisms and fungal virulence are strictly connected. Proteins of the endosomal sorting complex required for transport (ESCRT) have been recently associated with polysaccharide export in the yeast-like human pathogen Cryptococcus neoformans. Snf7 is a key ESCRT operator required for unconventional secretion in Eukaryotes. In this study we generated snf7Δ mutant strains of C. neoformans and its sibling species C. gattii. Lack of Snf7 resulted in important alterations in polysaccharide secretion, capsular formation and pigmentation. This phenotype culminated with loss of virulence in an intranasal model of murine infection in both species. Our data support the notion that Snf7 expression regulates virulence in C. neoformans and C. gattii by ablating polysaccharide and melanin traffic. These results are in agreement with the observation that unconventional secretion is essential for cryptococcal pathogenesis and strongly suggest the occurrence of still obscure mechanisms of exportation of non-protein molecules in Eukaryotes.
Similar content being viewed by others
Introduction
Cryptococcosis is the leading cause of human deaths related to fungal infections in Africa1. This disease is caused by the yeast-like pathogens Cryptococcus neoformans and C. gattii. The former is an opportunistic fungal pathogen that causes highly lethal meningitis in immunocompromised individuals2. C. gattii, on the other hand, can infect both immunocompromised and immunocompetent individuals3.
The progress of cryptococcal disease relies on several extracellular virulence factors4,5, implying that secretion is fundamental for cryptococcal pathogenesis. Eukaryotic molecules are exported through different cellular pathways, including conventional and unconventional secretory routes6,7. Conventionally secreted proteins follow the classical endoplasmic reticulum (ER)/Golgi-dependent secretory pathway7. Protein secretion through conventional mechanisms is dependent of an N-terminus-linked signal peptide, which is responsible for translocation of polypeptides into the lumen of the ER7. Proteins that lack a typical signal peptide are transported independently of this classical ER-Golgi route and engage the so-called unconventional secretion mechanisms6.
Most of the pathogenic determinants of C. neoformans lack secretory tags, supporting the notion that unconventional pathways of secretion are required for cryptococcal virulence8. C. neoformans and C. gattii export massive amounts of polysaccharides to the extracellular milieu9,10. Polysaccharide export is required for capsule formation, which is essential for the survival of cryptococci in host tissues11,12,13,14. The main component of the polysaccharide capsule is glucuronoxylomannan (GXM). GXM is co-transported to the extracellular space with other virulence factors inside vesicles that traverse the cell wall15,16,17. Although the morphological aspects of these vesicles suggest that they are similar to mammalian exosomes, their cellular origin remains undetermined (reviewed by Oliveira and colleagues18).
The eukaryotic sucrose non-fermenting protein 7 (Snf7) is a key operator in the endosomal sorting complex required for transport (ESCRT)-III19,20. This molecular complex, in association with other ESCRT components, is involved in multivesicular body (MVB) formation and cargo. MVB formation requires Snf7 oligomerization into a membrane-associated filament, which is nucleated by its association with the vacuolar protein sorting regulator Vps2021. MVBs, which derive from late endosomes, can be directed for fusion with the plasma membrane, resulting in extracellular release of their luminal vesicles, the so-called exosomes22,23. The ESCRT machinery has also been implicated in environmental pH sensing and adaptation in various fungal species through mechanisms that require the RIM101 pathway, a signaling mechanism necessary for the regulation of alkaline environment-related genes24,25. Recently, the RIM101-mediated stress response has been demonstrated to be up regulated during human meningitis caused by C. neoformans26. The supposition that MVBs are associated with the export of virulence factors in C. neoformans17 and the fact that adaptation to alkaline pHs is required for fungal survival in both alveolar spaces and the bloodstream27 suggests that SNF7 is involved in cryptococcal pathogenicity.
In this work we generated C. neoformans and C. gattiisnf7Δ mutant strains and characterized their biological and pathogenic properties. Lack of Snf7 affected key virulence determinants in both C. neoformans and C. gattii. This phenotype culminated with loss of virulence in an intranasal model of murine infection in both species. These results strongly support the hypothesis that unconventional secretion pathways are essential for the pathogenesis of C. neoformans and C. gattii.
Results
Snf7-related sequences in C. neoformans and C. gattii
A database search of the C. gattii genome (http://www.broadinstitute.org/annotation/genome/cryptococcus_neoformans_b/MultiHome.html- August 5, 2009, strain R265) revealed that the SNF7 (accession number CNBG_3856) coding region comprises 1,087 bp, includes five introns and encodes a protein of 221 amino acids. A similar search in the C. neoformans genome (http://www.broadinstitute.org/annotation/genome/cryptococcus_neoformans/MultiHome.html) identified the putative SNF7 sequence (accession number CNAG_01583.2) based on the similarity to the corresponding C. gattii sequence (accession number CNBG_3856). The C. neoformansSNF7 ortholog is 1,413 bp long, contains five introns and also encodes a putative 221 amino-acid protein. Phylogenetic analysis of Snf7 was performed with orthologues from distinct eukaryotic organisms (Figure 1A). The putative sequence of the C. gattii protein was highly similar to its C. neoformans counterpart (98% identity) and both display similarities to plant proteins (A. thaliana ~35% identity; O. sativa ~24% identity). In comparison to other fungi, the Snf7 sequences of both species studied here showed 45% identity with the S. cerevisiae protein and 43% identity with a C. albicans homologue.
Analysis of cryptococcal SNF7.
(A). Phylogenetic analysis was performed by applying the neighbor-joining method with Snf7 amino acid sequences from distinct organisms as follows: S. cerevisiae, C. albicans, A. nidulans, P. brasiliensis, S. pombe, N. crassa, M. oryzae, A. thaliana, O. sativa, C. neoformans and C. gattii. The bar marker indicates the genetic distance, which is proportional to the number of amino acid substitutions. Bootstrap values obtained with 1,000 re-samplings are displayed at the nodes. (B). Analysis of SNF7 disruption in C.neoformans and C. gattii by Southern blot. Genomic DNA (10 μg) from WT, snf7Δ and snf7Δ::SNF7 strains were digested with the Xbal restriction enzyme. The 3′ gene flank was used as probe in Southern hybridization. (C). For RT-PCR, cDNA from C. neoformans and C. gattii (WT, snf7Δ and snf7Δ::SNF7 strains) were used as template and genomic DNA of both fungi (WT cells) was used as positive control. Actin gene (ACT1) was used as reference gene for normalization of RT-PCR.
Disruption and complementation of SNF7 genes in both C. neoformans and C. gattii
To analyze the functions of SNF7, we generated snf7Δ disruption mutants in the backgrounds of the standard strains H99 (serotype A isolate of C. neoformans) and R265 (serotype B isolate of C. gattii). These mutant strains were called CN-snf7Δ and CG-snf7Δ, respectively. To ensure that the eventual phenotypes observed in the mutant strains were due to the knockout of the SNF7 gene, we constructed the complemented strains (CN-snf7Δ::SNF7 for C. neoformans and CG-snf7Δ::SNF7 for C. gattii) by integrating a wild-type copy of the SNF7 gene into the genome of the mutant strains. Deletion and complementation of SNF7 in both species were confirmed by Southern blot analysis (Figure 1B) and RT-PCR (Figure 1C).
Growth rates of the snf7Δ mutants under different conditions
We first evaluated whether the growth rates of the snf7Δ strains varied under different conditions. C. neoformans and C. gattii mutant strains showed normal growth at both 30 and 37°C, in comparison to parental and reconstituted strains (Figure 2A). Since copper acquisition is important for C. neoformans melanization and capsule formation28,29 we also evaluated the ability of the snf7Δ mutants to grow under copper deprivation conditions. Once again, the mutants showed normal growth rates, in comparison to WT and complemented cells (Figure 2B).
Effects of SNF7 disruption on fungal growth under different conditions.
(A). Wild type (WT), mutant (snf7Δ) and complemented (snf7Δ::SNF7) strains of C. neoformans and C. gattii were grown in YPD broth with shaking at 30°C or 37°C. Growth measurements (OD600) were performed at 4 h, 6 h, 8 h, 10 h, 12 h and 24 h. (B). Ten-fold serial dilutions of all strains were plated on LCM + Cu and LCM agar plates. The plates were incubated for 2 days at 30°C or 37°C. Fungal growth in LCM + Cu represented the control condition. (C). Growth rates of WT, mutant and complemented strains at pHs 7, 7.5, 8, 8.5 and 9. (D). Growth of WT, snf7Δ and snf7Δ::SNF7 strains on YPD agar plates supplemented with 150 mM LiCl2. Experiments were performed in triplicate and representative examples are shown.
The ESCRT machinery, including Snf7, plays a central role in resistance to higher pHs and tolerance to ionic lithium30,31,32. Therefore, we analyzed the ability of the snf7Δ mutants to survive under these conditions. Analysis of fungal growth in pHs ranging from 7 to 9 suggested lower growth rates of both CN-snf7Δ and CG-snf7Δ in pHs above 7.5, in comparison to parental and reconstituted cells (Figure 2C; p < 0.05 for all comparisons between cells expressing Snf7 and snf7Δ strains). A similar result was obtained when the C. neoformans and C. gattii strains were cultivated in the presence of 150 mM LiCl2 (Figure 2D). The phenotypes described above in C. neoformans and other fungal species are related to the interaction between Snf7 and molecules belonging to the RIM101 pathway (Figure 3A), which includes Rim101p and Rim20p30,33,34,35. Therefore, to support the notion that the phenotypes we observed were linked to defects in Rim101p activation via interaction with Snf7/Rim20p we assessed the expression of RIM101 and RIM20. We observed that although the mRNA levels of RIM20 remained the same for all strains, the expression of RIM101 was significantly lower in both snf7Δ mutants (Figure 3B and C). This observation is in agreement with a lower activation of RIM101p due to lack of Snf7.
SNF7 disruption affects the expression of RIM101.
(A). Conceptual basis for the analysis of Rim101 and Rim20 expression in C. neoformans and C. gattii strains. Rim101 activation depends on the recruitment of ESCRTI, ESCRTII and ESCRTIII components (Vps20 and Snf7) to the cytoplasmic face of the endosomal membrane. The Snf7 and Rim20/Rim13 protein complex interacts with the Rim101 precursor, which is further cleaved to release the active form of Rim101. This process results in the regulation of cryptococcal genes playing key roles in a number of cellular events. Adapted from Selvig and Alspaugh87. Expression levels of RIM101 and RIM20 were assessed in wild-type (WT), mutant (snf7) and complemented (snf7::SNF7) strains of C.neoformans (B) and C.gattii (C). The expression levels were normalized to the wild-type using the 2−ΔΔCt88.The experiments were performed with two biological samples and each cDNA sample was analyzed in triplicate with each primer pair. (*, p < 0.05 in comparison with WT and complemented cells).
SNF7 disruption alters pigmentation but not urease and phospholipase activities
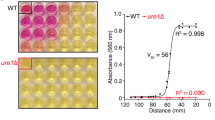
Extracellularly released enzymes and exported pigments are fundamental for cryptococcal pathogenesis5,17,36,37,38,39. Therefore, we analyzed whether SNF7 disruption would impact melanization and the extracellular activities of urease and phospholipase. Pigmentation was evaluated visually after growth of C. neoformans and C. gattii on both Niger seed and L-DOPA-containing solid media (Figure 4A). Under both conditions, the CN-snf7Δ mutant strain was similar to parental (WT) and complemented (reconstituted) strains in its ability to melanize at 30°C. However, when cultivated at 37°C, the mutant CN-snf7Δ showed reduced pigmentation. Differently, the CG-snf7Δ mutant showed subtle melanization defects when cultivated on L-DOPA agar at 30°C. At 37°C, the pigmentation defects were more evident. On Niger seed agar, SNF7 deletion in C. gattii affected pigmentation at 30°C, but not at 37°C. These results revealed an unexpected inter-species diversity in the relationship between Snf7 and pigmentation.
Disruption of SNF7 affects pigmentation but not extracellular enzyme activity.
(A). Fungal cells were grown on Niger seed agar and L-DOPA plates to evaluate melanin production by wild-type (WT), mutant (snf7Δ) and complemented (snf7Δ::SNF7) strains of C. neoformans and C. gattii. The cultures were incubated at 30°C and 37°C and monitored for 2 (Niger), 3 (L-DOPA 30°C) or 4 (L-DOPA 37°C) days of cultivation. (B). Phospholipase activity in egg yolk agar. All strains were inoculated on plates containing egg yolk agar and incubated at 30°C for 4 days and Pz values were then determined. (C). Urease activity levels of WT, snf7Δ, snf7Δ::SNF7 cells. Fungal cells were incubated in urea broth with shaking (200 rpm) at 37°C for 24 hours for colorimetric determination. Statistical differences were not observed when parental, mutant and complemented strains were compared.
Crude phospholipase activity was also assessed in the SNF7 disruption model. Although the rate of phospholipid hydrolysis showed some tendency to be higher in the CN-snf7Δ mutant, there were no statistical differences between any of the C. neoformans strains (Figure 4B). The C. gattii snf7 mutant showed levels of phospholipase activity that tended to be lower than those observed for parental and complemented strains (Figure 4B). Urease activity was similar in all strains analyzed in this study (Figure 4C).
Lack of Snf7 severely impairs GXM secretion and capsule formation
GXM secretion and capsule enlargement are essential for cryptococcal pathogenesis (reviewed in40). Since it has been suggested that GXM is exported by unconventional secretion mechanisms (reviewed elsewhere9,15,41), we evaluated whether SNF7 deletion affects capsule size and extracellular polysaccharide secretion.
Quantification of secreted GXM in all strains by ELISA revealed that deletion of SNF7 nearly extinguished polysaccharide export in both C. neoformans and C. gattii (Figure 5A). Morphological analysis of the capsule by India ink counterstaining, immunostaining of GXM and scanning electron microscopy revealed a clear reduction in capsular dimensions in the snf7Δ mutants (Figure 5B), implying defects in capsular assembly. Determination of the capsule/cell diameter ratio confirmed a significant reduction (p < 0.0001) in capsular dimensions in both CN-snf7Δ and CG-snf7Δ cells (Figure 5C).
Surface architecture and GXM release are affected by SNF7 deletion.
(A). Determination of GXM in C. neoformans and C. gattii culture supernatants. Asterisks denote statistically significant differences (**, p < 0.005; ***, p < 0.001) between the values obtained for the mutant strains and those obtained for wild type (WT) and complemented (snf7Δ::SNF7) cells. (B). Microscopic analysis of WT, snf7Δ and snf7Δ::SNF7 strains of C. neoformans and C. gattii. India ink counterstaining and immunostaining (upper and middle panels, respectively) revealed that SNF7 disruption profoundly affects capsule formation. GXM and chitin are stained in green and blue respectively (middle panels). Scale bars represent 5 μm. Scanning electron microscopy of the C. neoformans and C. gattii strains (Lower panels). Scale bar represents 1 μm. (C). Quantification of capsular dimensions based on in silico measurements of the results illustrated in B (***, p < 0.001).
snf7Δ mutants are avirulent in a murine infection model
The involvement of Snf7 in melanization, polysaccharide secretion and capsule assembly was in agreement with the possibility that SNF7 deletion could affect pathogenesis. To address this question, mice were infected intranasally with each strain used in this work for mortality assessment. The totality of animals infected with wild type and CN-snf7Δ::SNF7 reconstituted strains died by day 14 post infection (Figure 6A). Infection of mice with the CN-snf7Δ strain, however, resulted in nearly 63% survival at day 40-post infection, when animals were sacrificed. Animals infected with the C. gattiisnf7Δ mutant had a survival rate of 87.5% by day 40 (Figure 6B). Mice infected with parental and complemented strains of C. gattii died by days 18 and 29, respectively. Statistical analysis (Mantel-Cox test) confirmed that C. neoformans (p = 0.0065) and C. gattii (p = 0.0008) cells lacking Snf7 were less efficient than parental and complemented strains in killing mice.
Cryptococcal virulence is affected by SNF7 disruption.
Mortality rates of mice lethally infected with wild type (WT), mutant (snf7Δ) and complemented (snf7Δ::SNF7) strains of C. neoformans (A) and C. gattii (B) were monitored daily. SNF7 deletion was associated with a non-virulent phenotype in both cryptococcal species. Determination of GXM concentration (black bars) and lung CFU counts (grey bars) in the lungs of mice infected with wild type (CN-WT) and mutant (CN-snf7Δ) cells (C) revealed defective polysaccharide production and reduced survival of the mutant (**, p = 0.0017; ***, p < 0.0001).
To assess GXM secretion in vivo, mice were infected with C.neoformans for determination of fungal loads and polysaccharide concentration in lung tissues (Figure 6C). In pulmonary samples from infected mice with the CN-snf7Δ mutant, the concentration of extracellular GXM was nearly half the values obtained when mice were infected with parental cells (p = 0.0017). Fungal loads were also greatly reduced when animals were infected with the CN-snf7Δ mutant, in comparison to animals infected with the wild type strain (p < 0.0001).
Discussion
Secretory activity is mandatory for cryptococcal pathogenesis. The vast majority of the well-characterized molecular determinants of cryptococci are extracellular, including GXM, urease and phospholipase B17,37,42,43,44. In addition, C. neoformans melanin, which is also fundamental for virulence, is exported to the cell surface by vesicle-dependent secretory mechanisms17,45. Except for phospholipase B, all the above-mentioned virulence factors lack the leader peptide required for conventional secretion46. This observation and the recent notion that most fungal extracellular molecules lack secretory tags17 strongly suggest that unconventional secretion mechanisms and fungal virulence are strictly connected. Due to high efficacy of both C. neoformans and C. gattii in exporting massive amounts of polysaccharides, we used this model to investigate connections between unconventional export of molecules and fungal pathogenesis.
The ESCRT machinery participates in MVB formation (reviewed in23,47), which is required for exosome release to the extracellular space. Snf7, one of the components of the ESCRTIII protein complex, is responsible for the genesis of intraluminal vesicles of MVBs47. Therefore, considering that fungal cells might use exosome-like vesicles for the transfer of intracellularly synthesized polysaccharides to the extracellular milieu17, it seems reasonable to suppose that Snf7 is required for unconventional export of GXM in fungal cells. This hypothesis is supported by the fact that C. neoformanssec mutants, which have defects in post-Golgi, conventional secretory mechanisms, have normal capsules48,49. On the other hand, C. neoformans ESCRT-I mutants lacking expression of Vps23 manifested defective capsule formation and reduced virulence in a mouse model of cryptococcosis50.
Most of the studies on secretory mechanisms in eukaryotic cells were focused on protein trafficking17,48,51,52,53,54,55. The mechanism for export of molecules of a non-protein nature, including pigments and polysaccharides, are poorly known. In mammalian cells, MVBs have been correlated with the initial biogenesis of melanosomes56, implying connections between unconventional secretion and pigment traffic. In plant cells, endomembranes participate in polysaccharide traffic57, but the identity of the components necessary for the transport of cell wall enzymes and polysaccharides is not known. In this study, deletion of SNF7 resulted in reduced efficacies of pigmentation, GXM secretion and capsule formation in C. neoformans and C. gattii. These observations suggest that MVB formation and unconventional secretion mechanisms participate directly in the export of surface and/or extracellular molecules of non-protein nature. Although this hypothesis still needs experimental confirmation, it agrees with the observation that in C. neoformans Golgi reassembly and stacking protein (GRASP) and the Apt1 flippase, which are both unconventional secretion regulators, were required for polysaccharide, but not protein export58,59,60.
Melanin is essential for cryptococcal virulence61. In our study, the effects of SNF7 deletion on melanin formation varied depending on the species analyzed, temperature of growth and source of substrates for pigment synthesis. This observation efficiently illustrates the physiological diversity between the sibling species C. neoformans and C. gattii, which is likely linked to their well-known differences in pathogenic potential3. These results also exemplify the functional diversity of Snf7 in eukaryotes. In our model, fungal growth was only slightly affected in mutants cultivated at 37°C, but a previous study with S. cerevisiae lacking Snf7 demonstrated a clear correlation between carbon source and temperature-sensitive growth62, suggesting an important metabolic variation that is affected by both nutrient availability and temperature of growth. The mechanisms explaining the differential ability of C. neoformans and C. gattii Snf7 mutants to produce pigments at different temperatures in the presence of distinct melanization substrates are still unknown, but our results and the seminal observations in the S. cerevisiae model62 clearly support the notion that the physiological functions of Snf7 are regulated at multiple levels and result in variable fungal phenotypes. It is noteworthy that the enzymatic activity required for melanization can be also influenced by pH. Due to the putative roles of Snf7 in pH sensing21,30, we cannot rule out the hypothesis that defective melanin formation resulted from altered laccase activity in the two species analyzed in our study.
SNF7 deletion also interfered with tolerance to alkaline pH and high lithium chloride concentrations. This data is in agreement with previous literature on S. cerevisiae63, C. albicans25,64 and A. nidulans65 suggesting the participation of Snf7 in the RIM101 signaling cascade. The RIM family includes a sensor protein (RIM21p) and a catalytic complex (RIM20p in association with RIM13p) capable of inducing the proteolytic activation of RIM101p. The lack of Snf7 has been correlated with inability to activate the RIM101 transcription factor independently of the presence of RIM20p and RIM13p25,35,64. In fact, the active form of Rim101p supposedly modulates the expression of several genes involved in the adaptation to different environments66 and its own expression, which is in agreement with the reduction of RIM101 mRNA observed in our study.
O'Meara and collaborators have recently shown that Rim101p is also essential for capsule enlargement in C. neoformans35. Although a rim101Δ mutant had normal GXM secretion, several genes involved in cell wall-biosynthesis were affected by RIM101 deletion, which was likely related to a hypocapsular phenotype35. The mutant retained virulence in a murine intranasal infection model, suggesting that secretion of GXM could sufficiently impact host cells in favor of the pathogenic process35. The results obtained in our study are in agreement with this supposition. It is noteworthy, however, that the SNF7 disruption mutants generated in this work additionally had reduced capsules and low efficacy to melanize, which likely renders these cells more susceptible to the antimicrobial arsenal produced by the host. Future studies on how SNF7 and RIM101 impact immunomodulation are necessary for a broader understanding on how these cellular pathways interfere with the pathogenic process.
Our observations support the notion that Snf7 expression regulates virulence in both C. neoformans and C. gattii by reducing polysaccharide and melanin traffic. These results further support the observation that unconventional secretion is essential for cryptococcal pathogenesis18,58,67 and strongly suggests the occurrence of still obscure mechanisms of exportation of molecules of a non-protein nature in Eukaryotes.
Methods
Fungal strains, plasmids and media
The C. neoformans strain H99 (serotype A) and the C. gattii strain R265 (serotype B) were the recipients for construction of the mutant strains. Cells were maintained in YPD medium (yeast extract 1%, peptone 2%, glucose 2% and 1.5% agar). To evaluate the effects of Cu2+ deprivation on C. neoformans and C. gattii, cells were grown in 10 mL of YPD broth previous to a further cultivation in 100 mL of Yeast Nitrogen Base broth (YNB) (Becton, Dickinson and Company, Sparks, USA) for 24 hours at 37°C. The cells (5 × 107 cells.mL−1) were then transferred to 50 mL of limited Cu2+medium (LCM) and of Cu2+-replete medium (LCM + Cu) for 12 hours at 37°C with shaking (200 rpm). LCM is identical to a previously described low iron medium (LIM)68, except for supplementation with 100 μM bathocuproine disulphonate (BCS) (Sigma Chemical Co., Saint Louis, USA) and 100 μM FeHEDTA (Sigma Chemical Co., Saint Louis, USA). LCM + Cu was prepared based on the composition described for iron-replete medium (LIM + Fe)68,69. YPD plates with nourseothricin (100 μg/mL) were used to select C. neoformansSNF7 deletion transformants (CN-snf7Δ). YPD plates with hygromycin (200 μg/mL) were used to select C. neoformansSNF7 complementation transformants (CN-snf7Δ::SNF7). For C. gattii, YPD plates supplemented with hygromycin (200 μg/mL) were used to select theSNF7 deletion transformants (CG-snf7Δ). YPD plates with supplemented nourseothricin (100 μg/mL) were used to select complemented transformants (CG-snf7Δ::SNF7). Plasmid pJAF1570 was the source of a hygromycin resistance cassette and pAI471 was the source of a nourseothricin resistance cassette. Plasmids were maintained in Escherichia coli grown at 37°C for 18 hours in LB broth or agar supplemented with 50 μg/mL of kanamycin.
In silico analysis of C. neoformans and C. gattii SNF7 orthologs
The C. neoformans putative SNF7 gene sequence (Broad Institute accession number CNAG_01583.2) was identified by a BLAST search in the C. neoformans var. grubii strain H99 genomic database at the Broad Institute database by using the SNF7 sequence of C. gattii (Broad Institute accession number CNBG_3856). The putative amino acid sequences of Snf7 functional homologs in Saccharomyces cerevisiae (NCBI accession number P39929.1), Candida albicans (NCBI accession number EEQ43071.1), Aspergillus nidulans (Broad Institute accession number ANID_04240), Paracoccidioides brasiliensis (Broad Institute accession number PAAG_07890.1), Schizosaccharomyces pombe (NCBI accession number Q9P7F7), Neurospora crassa (Broad Institute accession number NCU01282.5), Magnaporthe oryzae (NCBI accession number EHA47375.1), Arabidopsis thaliana (NCBI accession number AEE85595.1), Oryza sativa (NCBI accession number AAT85290.1), C. neoformans (Broad Institute accession number CNAG_01583.2) and C. gattii (Broad Institute accession number CNBG_3856) were aligned using Clustal X272. Phylogenetic analysis was performed using Mega5 applying the neighbor-joining method73. Tree architecture was inferred from 1,000 bootstraps.
Disruption and complementation of SNF7
Delsgate methodology74,75,76 was used to construct the snf7Δ mutant strains. For the C. neoformans mutant, pDONRNAT vector was constructed as previously described76. The 5′ and 3′ SNF7 flanks (~700 bp) were PCR amplified and gel purified using the Illustra GFX PCR DNA and gel band purification kit (GE Healthcare, Fairfield, USA). pDONRNAT (~300 ng) plus 5′ and 3′ PCR products (~30 ng of each) were submitted to BP clonase reaction, according to manufacturer's instructions (Invitrogen, Carlsbad, USA). This reaction mixture was transformed into Escherichia coli OmniMAX 2-T177. The deletion construct was linearized by I-SceI enzymatic digestion and submitted to biolistic transformation78 in C. neoformans (H99 strain). The possible snf7Δ mutant colonies were screened by PCR and the deletion was confirmed by Southern blot analysis and reverse transcription-PCR (RT-PCR). For complementation of the SNF7 mutant, a genomic PCR fragment containing a wild type C. neoformansSNF7 gene was cloned into the SmaI site of pJAF15 plasmid. Biolistic transformation was performed to introduce the resultant vector into snf7Δ mutant strain. Genomic insertion was confirmed by Southern blot and RT-PCR.
The generation of the C. gattii mutant and complemented strains was performed as described above for C. neoformans with some modifications. For the mutant strain, a hygromicin resistance cassette was used to construct pDONRHYG, which was transformed into E. coli TG2 cells. The mutant strains were screened and confirmed as described above. For complementation, a genomic PCR fragment containing the wild-type C. gattiiSNF7 gene was cloned into the SmaI site of the pAI4 plasmid. Genomic insertion was confirmed via Southern blot and RT-PCR.
Primers used to construct and confirm mutant and complemented strains are listed in Table 1.
Phenotypic assays
For spot assays, wild type C. neoformans and C. gattii (CN-WT and CG-WT), CN-snf7Δ and CG-snf7Δ mutants and complemented (CN-snf7Δ::SNF7 and CG-snf7Δ::SNF7) strains were cultivated in YPD medium and serially diluted75. Diluted cells were spotted in LCM agar, LCM + Cu agar and 150 mM LiCl264 (Sigma, St. Louis, MO). Melanin production was visualized in cells spotted on Niger seed agar plates and on defined media containing 1 mM of L-DOPA79,80. For capsule analysis, WT, snf7Δ and complemented strains of C. neoformans and C. gattii (5 × 106 cells/mL) were incubated using six well culture plates in DMEM (Invitrogen, Carlsbad, CA) at 37°C and 5% CO2 for 48 hours. Capsule measurements were performed as previously described76. Staining of surface components (chitin and GXM) was performed as described elsewhere81. Briefly, fungal cells were fixed in 4% paraformaldehyde (Sigma, St. Louis, MO) for 1 hour followed by a incubation with phosphate-buffered saline (PBS) supplemented with 1% bovine serum albumin (Sigma, St. Louis, MO). Cell structures were then stained using calcofluor white (25 μM) (Sigma, St. Louis, MO) and 18B7 anti GXM monoclonal antibody (1 μg/mL) (kindly provided by Dr. Arturo Casadevall) followed by anti murine IgG Alexa Fluor 488 conjugated (Invitrogen, Carlsbad, CA). Images were acquired using an Axyoplan 2 microscope (Carl Zeiss, Germany). Cell surface structures were also observed by scanning electron microscopy (SEM) as described elsewhere67. Analysis of extracellular GXM was performed according to the method described by Casadevall and colleagues82, with minor modifications83. Phospholipase activity was determined by using egg yolk agar84,85. Approximately 107 cells of WT, snf7Δ and complemented strains of both C. neoformans and C. gattii were spotted onto the center of petri dishes containing egg yolk agar and incubated at 30°C. After 4 days, the phospholipase activity (Pz value) was determined as described by Price and colleagues84. To test growth in alkaline pH, YPD medium was buffered with 150 mM HEPES to pH 7, 7.5, 8, 8.5 and 9 with NaOH. Cultures were serially diluted in 96-well microtiter plates containing a pH-tested medium. After incubation at 30°C for 24 hours, absorbance measurements (OD600) were performed using a ELx80 Absorbance Microplate Reader (BioTek, Winooski, VT) and analyzed using GraphPad Prism software. Urease activity was evaluated as described by Kwon-Chung and colleagues86. The measurements (OD560) in urea broth were done at 2, 4 and 24 hours interval. All phenotypic assays were performed in triplicate.
Real time quantitative reverse transcription PCR (RT-qPCR) analysis
Cultures of WT, snf7Δ and snf7Δ::SNF7 strains of C. neoformans and C. gattii were grown overnight in YPD medium at 30 °C with shaking. RNA extraction and cDNA synthesis were performed as previously described75 for all strains in this study. The Applied Biosystems 7500 real-time PCR System (Life Technologies, Carlsbad, CA) was used for real time PCRs and the PCR cycling conditions as well as melting curve and relative expression determination were performed as described75. The experiments were performed using two biological samples and each cDNA sample was analyzed in triplicate with each primer pair. Actin cDNA levels were used to normalize each set of PCR experiments. The primer sequences used are listed in the Table 1.
In vivo assays
Mice were intraperitoneally anesthetized with 100 mg/kg ketamine and 16 mg/kg xylazine, followed by intranasal infection5 with WT, snf7Δ and complemented strains of C. neoformans and C. gattii. For each strain tested, eight female BALB/c mice (approximately 5 weeks old) were used. Fungal cells were first grown in YPD medium at 30°C, 200 RPM shaking, during 20 hours. The cells were washed twice and suspended in PBS solution. The infections were performed by inoculation of 107 yeast cells suspended in 50 μL PBS per animal. Infected mice were monitored daily. Mantel-Cox tests were performed using GraphPad Prism software. For CFU determination and in vivo GXM analysis, groups of 3 female BALB/c mice (approximately 5 weeks old) were inoculated intratracheally with 107 cells of CN-WT and CN-snfΔ strains. After 6 days animals were sacrificed and the lungs were removed and macerated in 5 mL YPD at 4°C. Macerates were clarified by centrifugation and supernatants were quantitatively assessed for the presence of GXM by ELISA as described previously82. Alternatively, 5 μL of the macerates were plated on YPD solid medium for further incubation at 37°C (48 h) and CFU counting.
Ethics Statement
The Universidade Federal do Rio Grande do Sul Ethics Committee for Use of Animals (CEUA - protocol number 19801) approved the use of animals in the present work, which were cared for according to the Brazilian National Council for Animal Experimentation Control (CONCEA) and Brazilian College of Animal Experimentation (COBEA) guidelines. Mice were kept in filtered top ventilated cages with food and water ad libitum. Groups of a maximum number of four mice were kept in each cage. All efforts to minimize animal suffering and to reduce the number of animals used were made. Animals were observed twice daily for any signals of suffering and/or for mortality during 40 days.
Statistical analysis
Statistical comparisons were performed with the Graphpad 5.0 software. Paired comparisons between different groups were performed using the Student's t test. Survival curves were statistically analyzed using the Mantel-Cox test.
References
Park, B. J. et al. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23, 525–30 (2009).
Chayakulkeeree, M. & Perfect, J. R. Cryptococcosis. Infect Dis Clin North Am 20, 507–44, v-vi (2006).
Byrnes, E. J., 3rd, Bartlett, K. H., Perfect, J. R. & Heitman, J. Cryptococcus gattii: an emerging fungal pathogen infecting humans and animals. Microbes Infect 13, 895–907 (2011).
Steenbergen, J. N. & Casadevall, A. The origin and maintenance of virulence for the human pathogenic fungus Cryptococcus neoformans. Microbes Infect 5, 667–75 (2003).
Cox, G. M., Mukherjee, J., Cole, G. T., Casadevall, A. & Perfect, J. R. Urease as a virulence factor in experimental cryptococcosis. Infect Immun 68, 443–8 (2000).
Nickel, W. & Rabouille, C. Mechanisms of regulated unconventional protein secretion. Nat Rev Mol Cell Biol 10, 148–55 (2009).
Schekman, R. Charting the secretory pathway in a simple eukaryote. Mol Biol Cell 21, 3781–4 (2010).
Rodrigues, M. L., Franzen, A. J., Nimrichter, L. & Miranda, K. Vesicular mechanisms of traffic of fungal molecules to the extracellular space. Curr Opin Microbiol 16, 414–20 (2013).
Casadevall, A., Nosanchuk, J. D., Williamson, P. & Rodrigues, M. L. Vesicular transport across the fungal cell wall. Trends Microbiol 17, 158–62 (2009).
Vecchiarelli, A. Immunoregulation by capsular components of Cryptococcus neoformans. Med Mycol 38, 407–17 (2000).
Bose, I., Reese, A. J., Ory, J. J., Janbon, G. & Doering, T. L. A yeast under cover: the capsule of Cryptococcus neoformans. Eukaryot Cell 2, 655–63 (2003).
Garcia-Rodas, R. & Zaragoza, O. Catch me if you can: phagocytosis and killing avoidance by Cryptococcus neoformans. FEMS Immunol Med Microbiol 64, 147–61 (2012).
Zaragoza, O. et al. The capsule of the fungal pathogen Cryptococcus neoformans. Adv Appl Microbiol 68, 133–216 (2009).
Villena, S. N. et al. Capsular polysaccharides galactoxylomannan and glucuronoxylomannan from Cryptococcus neoformans induce macrophage apoptosis mediated by Fas ligand. Cell Microbiol 10, 1274–85 (2008).
Rodrigues, M. L. et al. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot Cell 6, 48–59 (2007).
Rodrigues, M. L. & Djordjevic, J. T. Unravelling secretion in Cryptococcus neoformans: more than one way to skin a cat. Mycopathologia 173, 407–18 (2012).
Rodrigues, M. L. et al. Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryot Cell 7, 58–67 (2008).
Oliveira, D., Rizzo, J., Joffe, L., Godinho, R. & Rodrigues, M. Where Do They Come from and Where Do They Go: Candidates for Regulating Extracellular Vesicle Formation in Fungi. Int J Mol Sci 14, 9581–9603 (2013).
Weiss, P., Huppert, S. & Kolling, R. Analysis of the dual function of the ESCRT-III protein Snf7 in endocytic trafficking and in gene expression. Biochem J 424, 89–97 (2009).
Shestakova, A. et al. Assembly of the AAA ATPase Vps4 on ESCRT-III. Mol Biol Cell 21, 1059–71 (2010).
Peck, J. W., Bowden, E. T. & Burbelo, P. D. Structure and function of human Vps20 and Snf7 proteins. Biochem J 377, 693–700 (2004).
Michelet, X., Djeddi, A. & Legouis, R. Developmental and cellular functions of the ESCRT machinery in pluricellular organisms. Biol Cell 102, 191–202 (2010).
Babst, M. MVB vesicle formation: ESCRT-dependent, ESCRT-independent and everything in between. Curr Opin Cell Biol 23, 452–7 (2011).
Maeda, T. The signaling mechanism of ambient pH sensing and adaptation in yeast and fungi. FEBS J 279, 1407–13 (2012).
Wolf, J. M. & Davis, D. A. Mutational analysis of Candida albicans SNF7 reveals genetically separable Rim101 and ESCRT functions and demonstrates divergence in bro1-domain protein interactions. Genetics 184, 673–94 (2010).
Chen, Y. et al. The Cryptococcus neoformans Transcriptome at the Site of Human Meningitis. MBio 5, e01087–13 (2014).
Rittershaus, P. C. et al. Glucosylceramide synthase is an essential regulator of pathogenicity of Cryptococcus neoformans. J Clin Invest 116, 1651–9 (2006).
Chun, C. D. & Madhani, H. D. Ctr2 links copper homeostasis to polysaccharide capsule formation and phagocytosis inhibition in the human fungal pathogen Cryptococcus neoformans. PLoS One 5, e12503 (2010).
Silva, F. D. et al. Effects of microplusin, a copper-chelating antimicrobial peptide, against Cryptococcus neoformans. FEMS Microbiol Lett 324, 64–72 (2011).
Kullas, A. L., Li, M. & Davis, D. A. Snf7p, a component of the ESCRT-III protein complex, is an upstream member of the RIM101 pathway in Candida albicans. Eukaryot Cell 3, 1609–18 (2004).
Wolf, J. M. & Davis, D. A. Mutational analysis of Candida albicans SNF7 reveals genetically separable Rim101 and ESCRT functions and demonstrates divergence in bro1-domain protein interactions. Genetics 184, 673–94 (2010).
Xu, W., Smith, F. J., Jr, Subaran, R. & Mitchell, A. P. Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans. Mol Biol Cell 15, 5528–37 (2004).
Bowers, K. et al. Protein-protein interactions of ESCRT complexes in the yeast Saccharomyces cerevisiae. Traffic 5, 194–210 (2004).
Lamb, T. M., Xu, W., Diamond, A. & Mitchell, A. P. Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J Biol Chem 276, 1850–6 (2001).
O'Meara, T. R. et al. Interaction of Cryptococcus neoformans Rim101 and protein kinase A regulates capsule. PLoS Pathog 6, e1000776 (2010).
Chen, S. C. et al. Identification of extracellular phospholipase B, lysophospholipase and acyltransferase produced by Cryptococcus neoformans. Infect Immun 65, 405–11 (1997).
Cox, G. M. et al. Extracellular phospholipase activity is a virulence factor for Cryptococcus neoformans. Mol Microbiol 39, 166–75 (2001).
Park, M., Do, E. & Jung, W. H. Lipolytic enzymes involved in the virulence of human pathogenic fungi. Mycobiology 41, 67–72 (2013).
Kronstad, J. W. et al. Expanding fungal pathogenesis: Cryptococcus breaks out of the opportunistic box. Nat Rev Microbiol 9, 193–203 (2011).
O'Meara, T. R. & Alspaugh, J. A. The Cryptococcus neoformans capsule: a sword and a shield. Clin Microbiol Rev 25, 387–408 (2012).
Rodrigues, M. L. et al. Vesicular transport systems in fungi. Future Microbiol 6, 1371–81 (2011).
Noverr, M. C., Cox, G. M., Perfect, J. R. & Huffnagle, G. B. Role of PLB1 in pulmonary inflammation and cryptococcal eicosanoid production. Infect Immun 71, 1538–47 (2003).
Santangelo, R. et al. Role of extracellular phospholipases and mononuclear phagocytes in dissemination of cryptococcosis in a murine model. Infect Immun 72, 2229–39 (2004).
Wright, L. C. et al. Cryptococcal phospholipases: a novel lysophospholipase discovered in the pathogenic fungus Cryptococcus gattii. Biochem J 384, 377–84 (2004).
Eisenman, H. C., Frases, S., Nicola, A. M., Rodrigues, M. L. & Casadevall, A. Vesicle-associated melanization in Cryptococcus neoformans. Microbiology 155, 3860–7 (2009).
Turner, K. M., Wright, L. C., Sorrell, T. C. & Djordjevic, J. T. N-linked glycosylation sites affect secretion of cryptococcal phospholipase B1, irrespective of glycosylphosphatidylinositol anchoring. Biochim Biophys Acta 1760, 1569–79 (2006).
Hanson, P. I. & Cashikar, A. Multivesicular body morphogenesis. Annu Rev Cell Dev Biol 28, 337–62 (2012).
Panepinto, J. et al. Sec6-dependent sorting of fungal extracellular exosomes and laccase of Cryptococcus neoformans. Mol Microbiol 71, 1165–76 (2009).
Yoneda, A. & Doering, T. L. A eukaryotic capsular polysaccharide is synthesized intracellularly and secreted via exocytosis. Mol Biol Cell 17, 5131–40 (2006).
Hu, G. et al. Cryptococcus neoformans requires the ESCRT protein Vps23 for iron acquisition from heme, for capsule formation and for virulence. Infect Immun 81, 292–302 (2013).
Oliveira, D. L. et al. Characterization of yeast extracellular vesicles: evidence for the participation of different pathways of cellular traffic in vesicle biogenesis. PLoS One 5, e11113 (2010).
Albuquerque, P. C. et al. Vesicular transport in Histoplasma capsulatum: an effective mechanism for trans-cell wall transfer of proteins and lipids in ascomycetes. Cell Microbiol 10, 1695–710 (2008).
Duran, J. M., Anjard, C., Stefan, C., Loomis, W. F. & Malhotra, V. Unconventional secretion of Acb1 is mediated by autophagosomes. J Cell Biol 188, 527–36 (2010).
Manjithaya, R., Anjard, C., Loomis, W. F. & Subramani, S. Unconventional secretion of Pichia pastoris Acb1 is dependent on GRASP protein, peroxisomal functions and autophagosome formation. J Cell Biol 188, 537–46 (2010).
Oliveira, D. L. et al. Biogenesis of extracellular vesicles in yeast: Many questions with few answers. Commun Integr Biol 3, 533–5 (2010).
Burgoyne, T. et al. Expression of OA1 limits the fusion of a subset of MVBs with lysosomes - a mechanism potentially involved in the initial biogenesis of melanosomes. J Cell Sci 126, 5143–52 (2013).
Kim, S. J. & Brandizzi, F. The Plant Secretory Pathway: An Essential Factory for Building the Plant Cell Wall. Plant Cell Physiol 55, 687–693 (2014).
Kmetzsch, L. et al. Role for Golgi reassembly and stacking protein (GRASP) in polysaccharide secretion and fungal virulence. Mol Microbiol 81, 206–18 (2011).
Hu, G. & Kronstad, J. W. A putative P-type ATPase, Apt1, is involved in stress tolerance and virulence in Cryptococcus neoformans. Eukaryot Cell 9, 74–83 (2010).
Rizzo, J. et al. Role of the Apt1 Protein in Polysaccharide Secretion by Cryptococcus neoformans. Eukaryot Cell 13, 715–26 (2014).
Nosanchuk, J. D. & Casadevall, A. Impact of melanin on microbial virulence and clinical resistance to antimicrobial compounds. Antimicrob Agents Chemother 50, 3519–28 (2006).
Tu, J., Vallier, L. G. & Carlson, M. Molecular and genetic analysis of the SNF7 gene in Saccharomyces cerevisiae. Genetics 135, 17–23 (1993).
Xu, W., Smith, F. J., Subaran, R. & Mitchell, A. P. Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans. Mol Biol Cell 15, 5528–37 (2004).
Kullas, A. L., Li, M. & Davis, D. A. Snf7p, a component of the ESCRT-III protein complex, is an upstream member of the RIM101 pathway in Candida albicans. Eukaryot Cell 3, 1609–18 (2004).
Tilburn, J., Sánchez-Ferrero, J. C., Reoyo, E., Arst, H. N. & Peñalva, M. A. Mutational analysis of the pH signal transduction component PalC of Aspergillus nidulans supports distant similarity to BRO1 domain family members. Genetics 171, 393–401 (2005).
Peñalva, M. A. & Arst, H. N. Regulation of gene expression by ambient pH in filamentous fungi and yeasts. Microbiol Mol Biol Rev 66, 426–46, table of contents (2002).
Rizzo, J. et al. Role of the Apt1 protein in polysaccharide secretion by Cryptococcus neoformans. Eukaryot Cell 13, 715–726 (2014).
Jacobson, E. S., Goodner, A. P. & Nyhus, K. J. Ferrous iron uptake in Cryptococcus neoformans. Infect Immun 66, 4169–75 (1998).
Lian, T. et al. Iron-regulated transcription and capsule formation in the fungal pathogen Cryptococcus neoformans. Mol Microbiol 55, 1452–72 (2005).
Fraser, J. A., Subaran, R. L., Nichols, C. B. & Heitman, J. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot Cell 2, 1036–45 (2003).
Idnurm, A., Reedy, J. L., Nussbaum, J. C. & Heitman, J. Cryptococcus neoformans virulence gene discovery through insertional mutagenesis. Eukaryot Cell 3, 420–9 (2004).
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–8 (2007).
Tamura, K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol Biol Evol 28, 2731–9 (2011).
García-Pedrajas, M. D. et al. DelsGate, a robust and rapid gene deletion construction method. Fungal Genet Biol 45, 379–88 (2008).
Kmetzsch, L. et al. The vacuolar Ca(2)(+) exchanger Vcx1 is involved in calcineurin-dependent Ca(2)(+) tolerance and virulence in Cryptococcus neoformans. in Eukaryot Cell, Vol. 9, 1798–805 (2010).
Kmetzsch, L. et al. The GATA-type transcriptional activator Gat1 regulates nitrogen uptake and metabolism in the human pathogen Cryptococcus neoformans. Fungal Genet Biol 48, 192–9 (2011).
Schneider Rde, O. et al. Zap1 Regulates Zinc Homeostasis and Modulates Virulence in Cryptococcus gattii. PLoS One 7, e43773 (2012).
Toffaletti, D. L., Rude, T. H., Johnston, S. A., Durack, D. T. & Perfect, J. R. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J Bacteriol 175, 1405–11 (1993).
Kmetzsch, L. et al. Role for Golgi reassembly and stacking protein (GRASP) in polysaccharide secretion and fungal virulence. Mol Microbiol 81, 206–18 (2011).
Baker, L. G., Specht, C. A., Donlin, M. J. & Lodge, J. K. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryot Cell 6, 855–67 (2007).
Rodrigues, M. L., Alvarez, M., Fonseca, F. L. & Casadevall, A. Binding of the wheat germ lectin to Cryptococcus neoformans suggests an association of chitinlike structures with yeast budding and capsular glucuronoxylomannan. Eukaryot Cell 7, 602–9 (2008).
Casadevall, A., Mukherjee, J. & Scharff, M. D. Monoclonal antibody based ELISAs for cryptococcal polysaccharide. J Immunol Methods 154, 27–35 (1992).
Fonseca, F. L. et al. Structural and functional properties of the Trichosporon asahii glucuronoxylomannan. Fungal Genet Biol 46, 496–505 (2009).
Price, M. F., Wilkinson, I. D. & Gentry, L. O. Plate method for detection of phospholipase activity in Candida albicans. Sabouraudia 20, 7–14 (1982).
Chen, S. C., Muller, M., Zhou, J. Z., Wright, L. C. & Sorrell, T. C. Phospholipase activity in Cryptococcus neoformans: a new virulence factor? J Infect Dis 175, 414–20 (1997).
Kwon-Chung, K. J., Wickes, B. L., Booth, J. L., Vishniac, H. S. & Bennett, J. E. Urease inhibition by EDTA in the two varieties of Cryptococcus neoformans. Infect Immun 55, 1751–4 (1987).
Selvig, K. & Alspaugh, J. A. pH Response Pathways in Fungi: Adapting to Host-derived and Environmental Signals. Mycobiology 39, 249–56 (2011).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–8 (2001).
Acknowledgements
We thank Fernanda L. Fonseca for help with microscopy tests and Debora L. Oliveira for help with in vivo experimentation. We are also grateful to Drs. Josh Nosanchuk, Maurizio Del Poeta and Leonardo Nimrichter for helpful suggestions. This work was supported by grants from the Brazilian agencies CNPq, CAPES, FAPERJ and MCT/FINEP/Rede GENOPROT (grant number 01.07.0552.00). JC received a scholarship (grant number 201227/2009-6) for a 6 months stay at The Scripps Research Institute, USA. The authors also acknowledge support from the Instituto Nacional de Ciência e Tecnologia de Inovaça~o em Doenças Negligenciadas (INCT-IDN).
Author information
Authors and Affiliations
Contributions
R.M.C.G., J.C., M.H.V. and M.L.R. wrote the main manuscript text; R.M.C.G. and J.C. prepared all figures; R.M.C.G., J.C., G.S.A., S.F. and L.K. conducted the experiments. R.M.C.G., J.C., G.S.A., S.F., L.K., C.C.S., A.S., M.H.V. and M.L.R. analyzed the data and reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
da C. Godinho, R., Crestani, J., Kmetzsch, L. et al. The vacuolar-sorting protein Snf7 is required for export of virulence determinants in members of the Cryptococcus neoformans complex.. Sci Rep 4, 6198 (2014). https://doi.org/10.1038/srep06198
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep06198
This article is cited by
-
Evaluation of reference genes for real-time quantitative PCR analysis in southern corn rootworm, Diabrotica undecimpunctata howardi (Barber)
Scientific Reports (2019)
-
Deciphering Fungal Extracellular Vesicles: From Cell Biology to Pathogenesis
Current Clinical Microbiology Reports (2019)
-
Systematic functional analysis of kinases in the fungal pathogen Cryptococcus neoformans
Nature Communications (2016)
-
Pathogenic diversity amongst serotype C VGIII and VGIV Cryptococcus gattii isolates
Scientific Reports (2015)