Abstract
Cutibacterium acnes (formerly Propionibacterium acnes) is recognized as a pathogen in foreign-body infections (arthroplasty or spinal instrumentation). To date, the direct impact of C. acnes on bone cells has never been explored. The clade of 11 C. acnes clinical isolates was determined by MLST. Human osteoblasts and osteoclasts were infected by live C. acnes. The whole genome sequence of six isolates of this collection was analyzed. CC36 C. acnes strains were significantly less internalized by osteoblasts and osteoclasts than CC18 and CC28 C. acnes strains (p ≤ 0.05). The CC18 C. acnes ATCC6919 isolate could survive intracellularly for at least 96 hours. C. acnes significantly decreased the resorption ability of osteoclasts with a major impact by the CC36 strain (p ≤ 0.05). Genome analysis revealed 27 genes possibly linked to these phenotypic behaviors. We showed a direct impact of C. acnes on bone cells, providing new explanations about the development of C. acnes foreign-body infections.
Similar content being viewed by others
Introduction
Bone is a mineralized tissue constantly remodeled under the simultaneous coordinated action of three cell types: bone matrix-resorbing osteoclasts, bone matrix-forming osteoblasts, and osteocytes embedded in the mineralized bone matrix1. This physiological process is tightly regulated and crucial to maintain a constant bone mass in adults. However, this balance can be impaired to favor resorption in pathological conditions, such as osteoporosis or bone and joint infection (BJI). The pathogenicity of Cutibacterium acnes has long been restricted to skin conditions2. However, C. acnes is increasingly recognized as a pathogen, mainly in foreign-body infections, such as arthroplasty or spinal instrumentation-associated infections3,4. The role of C. acnes in foreign-body infections is probably underestimated because, as an anaerobe, it needs an adapted bacterial transport system and usually a prolonged incubation time of up to 14 days for growth5. Late growth and/or growth only in enrichment media are often misinterpreted as contamination6. Recognition of the pathogenicity of C. acnes in such cases is dependent on its recovery in multiple, separately collected perioperative samples from a site expected to be sterile, in association with clinical symptoms7. Unfortunately, the clinical diagnosis of such low-grade infection can be challenging because these infections often mimic aseptic loosening with scarce or nonspecific clinical signs and symptoms8. The absence of a gold standard for the diagnosis and accurate criteria to define foreign-body infection may explain why C. acnes torpid infections are sometimes misclassified as aseptic loosening or mechanical failure9.
Therefore, the association between implant surgery and C. acnes infection is not always obvious10 and different studies have focused on population genetics and sequence typing in an attempt to identify sub-populations with different pathogenic potential11,12,13,14,15,16. To date, no clear association between phylotypes and infection/colonization has been reported. A genetic analysis of 210 C. acnes isolates from various origins demonstrated that phylotypes IB, II and III were more often associated with blood, cerebrospinal fluid and prosthetic hip infections whereas phylotype IA strains were highly associated with acne13. IB strains have also been more frequently isolated from infected prostheses than IA strains 17. Although several typing methods have been developed for C. acnes, whole genome-based typing reveals that only MLST methods have the discriminatory power to identify all distinct subtypes13. Recently, Yu et al. stated that studies focusing on well characterized types may further increase our understanding of the role of different C. acnes types in the pathogenesis of disease18.
Based on this statement, and because the direct impact of C. acnes isolates on bone cells has never been explored, we investigated the sequence type (ST) of C. acnes clinical isolates and developed different models in order to study the interaction between C. acnes and bone cells (osteoblasts or osteoclasts). Finally, whole genome sequencing (WGS) analysis revealed genetic tracks to explain the C. acnes behaviors observed.
Material and Methods
Bacterial strains
Eleven C. acnes clinical isolates (11 patients) were selected from a large collection of strains, including the main phylotypes and clonal complexes, mostly from previous studies19,20. Four isolates were recovered from monomicrobial spinal instrumentation infections, two from prosthetic monomicrobial infections, three from acne lesions and two were reference strains (ATCC11827 and ATCC6919). The characteristics of the selected isolates are summarized in Table 1. Implant-associated infections were confirmed using the Infectious Diseases Society of America criteria for bone and joint infections21.
Molecular methods
Clonality
Multi-Locus Sequence Typing (MLST) was carried out on all isolates as described by Lomholt et al.13. This scheme is based on partial sequences of nine housekeeping genes comprising a total of 4,287 nucleotides and is available at http://pacnes.mlst.net/.
Whole genome sequencing
To investigate the relationship between phenotypic and genotypic results, WGS was performed on a subset from our collection. Five isolates were selected depending on their sequence type and clinical origin (Table 1). Bacterial DNA was extracted according to the manufacturer’s protocol using the commercial extraction kit QIAamp® DNA mini kit (Qiagen, Hilden, Germany). Genome sequencing was performed using the MiSeq (Illumina) sequencing technology22. The genome alignment of C. acnes isolates was visualized using a progressive Mauve algorithm in the Mauve program version 20150226 build 1023. C. acnes genome sequences were annotated using the Rapid Annotation Subsystem Technology (RAST) automated web service24. Figure for genome comparison was generated by BRIG software25 and OAT software26.
Co-culture with bone cells
Before infection of the bone cell cultures, strains were grown overnight at 37 °C in 40 mL of Brain Heart Infusion (BHI, Oxoid, Dardilly, France), harvested by centrifugation for 10 min at 800 g and washed once in 5 mL of phosphate-buffered saline (PBS). The pellets were suspended in 5 mL of Dulbecco’s Modified Eagle’s Medium (DMEM; Invitrogen-Life Technologies, Inc.). Bacterial suspensions were adjusted with a nephelometer to obtain 1 × 108 CFU mL−1 and then diluted in DMEM.
Human osteoblast cultures
The human osteosarcoma cell line MG-63 purchased from ATCC was cultured in a 5% CO2 atmosphere at 37 °C in DMEM supplemented with 5% of fetal bovine serum (Hyclone Perbio, France). Three days before infection, cells were seeded in 24-well culture plates to obtain confluent monolayers. To confirm the results obtained with MG-63 cells, the same method was applied with two representative isolates (ATCC6919 and BL strains) on human mesenchymal stem cells (hMSC). hMSC were obtained from healthy donors. Blood draws were performed at the “Etablissement Français du Sang” (Nantes, France) after obtaining the informed consent of all healthy donors. The experimental protocol was approved by the ethics committee of the Medical School of Nantes. All experiments were performed in accordance with the Good Scientific Practice guidelines of the Medical School of Nantes and all relevant guidelines and regulations. hMSC were cultured in DMEM supplemented with 10% of fetal bovine serum, 1 ng/mL of basic Fibroblast Growth Factor (bFGF; R&D systems, UK), 100 U/mL of penicillin/streptomycin, and 2 mM L-glutamine. Adherent cells were frozen at passage 2 after characterization by flow cytometry (CD45−, CD34−, CD105+, CD73+, and CD90+, purity ≥99%) prior to further experiments27.
Internalization assay
Bacterial internalization experiments were adapted from Crémet et al. study28. Briefly, the MG-63 monolayers were infected for 2 h (multiplicity of infection (MOI) of 100:1). The number of bacteria present in the inocula was checked by plating dilutions on Schaedler plates incubated under anaerobic conditions. After 2 h of infection, cells were washed twice with PBS and incubated for a further 2 h in 1 mL of DMEM containing 1% of penicillin/streptomycin to kill the remaining extracellular bacteria. The cell monolayers were then washed three times and lysed with 0.1% Triton X-100. The lysates were serially diluted and plated on Schaedler plates to count the number of intracellular bacteria. The results were expressed as percentages of inoculum.
Bacterial internalization assessment by cell staining and confocal microscopy
To confirm the bacterial internalization, MG-63 cells were seeded onto slides after the internalization assay and then Gram and Giemsa stained. For live confocal microscopy, MG-63 cells were seeded onto Ibidi® μ-Slide 18 well (Munich, Germany) and infected with two selected clinical strains (ATCC6919 and BL), as described above. Fluorescein isothiocyanate (Sigma-Aldrich®, Saint Quentin Fallavier, France) was used to visualize the bacteria, along with calcein red/orange (Thermo Fischer Scientific, Waltham, MA USA) to label the cellular membrane. Slides were acquired on a Nikon A1 Rsi confocal microscope designed for live cell imaging using an Okolab environmental chamber to regulate temperature and air/CO2/N2. Pictures were analyzed with Fiji software29. 3D images were processed with NIS elements (Nikon Instruments Inc.) and Volocity 3D Image Analysis Software (PerkinElmer).
Persistence assay
ATCC6919 and BL C. acnes strains were internalized in MG-63 cells as described above. Following incubation with penicillin/streptomycin, MG-63 cells were lysed at 12, 24, 48, 72, 96 and 168 h post-infection to assess intracellular bacterial viability and persistence. Lysates were seeded on Schaedler plates. Bacterial growth was observed after five days of incubation at 37 °C under anaerobic atmosphere conditions.
Cell viability and osteoclastogenesis assay
The generation of osteoclasts from human CD14+ monocytes has been described previously30. Briefly, purified CD14+ cells from different healthy donors were cultured in α-MEM with 10% FCS and 25 ng/mL human M-CSF (R&D systems). Blood draws were performed at the “Etablissement Français du Sang” (Nantes, France) after obtaining the informed consent of all healthy donors. The experimental protocol was approved by the ethics committee of the Medical School of Nantes. All experiments were performed in accordance with the Good Scientific Practice guidelines of the Medical School of Nantes and all relevant guidelines and regulations. At day 3, cells were infected with C. acnes as described above (MOI 10:1), followed by antibiotic treatment for 2 h and addition of 100 ng/mL RANKL. Multinucleated cells formed with 3 or more nuclei were counted after TRAP staining (Sigma, France). Cell growth was measured using the crystal violet assay as described previously30.
Bone resorption assay
Mature osteoclasts were obtained by differentiation of CD14+ cells and then infected by C. acnes as described above (MOI 10:1), followed by antibiotic treatment for 2 h. Next, cells were incubated for 48 h on 96-well Corning Osteo Assay® plates (OsteoCorning, Corning, MA). After the culture period, cells were removed with bleach solution and the area of resorption pits was quantified using Image J software (NIH, Bethesda, MD).
Results
C. acnes belonging to clonal complex 36 are less internalized by human osteoblast-like cells
The bacterial internalization assay by osteoblasts was carried out using 11 C. acnes strains. Clonal complexes of these isolates are shown in Table 1. CC36 C. acnes strains were less invasive than CC18 and CC28 C. acnes strains toward osteoblasts (mean percentage of internalized bacteria <0.01% for CC36 P. acnes strains versus more than 1% for CC18 and CC28 C. acnes strains (Fig. 1A)). The ATCC11827 CC18 C. acnes strain exhibited a similar invasiveness to CC36 isolates (Fig. 1A). To assess whether the observed results were not related to the nature of osteosarcoma MG-63 cells, we confirmed our results using mesenchymal stem cells. These had the advantage of sharing common features with all bone cells and enabled the results observed to be extrapolated to all derivative cells. The mean percentages obtained previously were similar for both isolates: CC18 ATCC6919 and CC36 BL C. acnes (Fig. 1B). Using these two isolates, the same results were also observed for the interaction, especially with osteoclasts (Fig. 1B). Determination of bacterial subcellular localization by Gram staining (Fig. 1C) and confocal microscopy (after stringent washing to remove unattached bacteria) confirmed the intracellular presence of C. acnes and the discrepancies observed between the clonal complexes (Fig. 1D,E, Supplementary data video 1).
(A) MG-63 cells were infected with different C. acnes strains at a MOI of 100:1 for 2 hours. After 2 h, cells were washed, treated with penicillin/streptomycin, washed again and lysed. CFU counts were determined after 5 days of anaerobic culture of the lysate on Schaedler plates. *P < 0.01. (B) MG-63 cells, osteoclasts (OC) and mesenchymal stem cells (MSC) were infected with different C. acnes strain at a MOI of 100:1 for 2 hours. After 2 h, cells were washed, treated with penicillin/streptomycin, washed again and lysed. CFU counts were determined after 5 days of anaerobic culture of the lysate on Schaedler plates. **P < 0.01. (C) MG-63 cells Gram stained (ATCC6919 C. acnes cells appear in purple). (D) Live confocal microscopy pictures after 2 h of incubation with MG-63 cells and washing. MG-63 cell membranes were stained with calcein red/orange (red). C. acnes bacteria were stained with fluorescein isothiocyanate (green). D1. Result with C. acnes ATCC6919 strain. D2. Result with C. acnes BL isolate. (E) Orthogonal views from different planes (x/y, x/z or y/z) of the live confocal microscopy images used to analyze the ATCC6919 CC18 C. acnes location. Bacteria (FITC, green) inside the cell (cellular membranes, stained with calcein red/orange, appear in red) are indicated by a white arrow.
C. acnes can survive in human osteoblast cells
Low-grade and chronic infections can be due to bacteria hidden from the immune response, inside cells. Herein, as C. acnes isolates demonstrated different osteoblast internalization abilities, we further evaluated whether CC18 ATCC6919 and CC36 BL C. acnes strains underwent clearance by osteoblasts. As shown in Fig. 2, the CC18 ATCC6919 strain could survive in osteoblasts after 96 h of incubation. A significant decrease in the number of viable intracellular bacteria was observed during the course of the experiment (p = 0.0007). The mean CFU/well ranged from 1.15 × 104 at time 0 to 17.3 after 96 h of incubation. No colony remained at Day 7. In contrast, no colony remained at 24 h with the CC36 BL strain. These results demonstrate the ability of C. acnes strains to remain viable intracellularly at least 96 h after infection, in our experimental model.
C. acnes infection has no impact on osteoclastogenesis
We then evaluated whether bacterial infection of the precursors could impact osteoclastogenesis. Cell viability was not significantly changed in any of the subgroups (Fig. 3A). Human CD14+ monocytes were cultured in control conditions or in the presence of ATCC6919 or CC36 BL C. acnes for 2 h. Seven days after internalization of ATCC6919 or CC36 BL C. acnes, a mean (±SD) of 382 ± 100 and 290 ± 6 of the cells, respectively, were multinucleated and TRAP-positive, compared with uninfected cells (416 ± 67) (Fig. 3B).
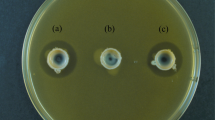
(A) Cell viability after infection of human CD14+ monocytes by C. acnes. NS: not significant. (B) Effect of infection of human CD14+ monocytes by C. acnes on osteoclastogenesis. Multinucleated cells formed with 3 or more nuclei (osteoclasts) were counted after TRAP staining. NS: not significant. (C) Resorption pit area due to mature osteoclasts uninfected (Ca), infected by C. acnes ATCC6919 (Cb), and infected by C. acnes BL (Cc). (D) Percentage of matrix resorbed by mature osteoclasts uninfected (control), or infected by C. acnes after 48 hours of culture. *P < 0.05 (one-tailed t-test).
C. acnes strains inhibit osteoclast bone resorption capacities
We then decided to evaluate the impact of C. acnes infection on mature osteoclast function. The mean percentage of the resorbed area (±SD) by cells infected with CC18 ATCC6919 or CC36 BL C. acnes strains was 9.6 ± 0.53% and 7 ± 1.9%, respectively. A significant difference (p < 0.05) was observed between uninfected wells (12 ± 1.7%) and C. acnes infected wells after 48 h of resorption. Moreover, CC36 C. acnes showed a more significant ability to decrease the resorption ability of osteoclasts than the CC18 ATCC6919 strain (p < 0.05). The results of the most representative experiment are presented in Fig. 3C,D.
Whole genome sequencing
WGS was performed on a subset of isolates to highlight candidate genes potentially involved in the differences in phenotypic behavior observed previously (Table 1). The ATCC6919, BL, Ntes, HB, 2003-1719 and 2004-10708 strains yielded 2617, 2469, 2416, 2410, 2463 and 2410 predicted coding sequences (CDSs), respectively31,32. Slightly less than half (44 to 47%) of the CDSs in each strain could be functionally categorized into 315 to 331 subsystems. Among the subsystems, carbohydrates, “cofactors, vitamins, prosthetic groups, pigments”, “amino acids and derivatives” and protein metabolism had the greatest number of genes. An overview of the genome characteristics of the isolates and their subsystem statistics is shown in Table 1 of the supplementary data. The SEED viewer identified 27 genes that were specific to the non-internalized isolates and 6 genes specific to the internalized isolates (Fig. 4, Table 2 in the supplementary data).
(A) General comparison of the whole genome sequence of C. acnes generated with OrthoANI values calculated from OAT (Orthologous Average Nucleotide Identity Tool) software (http://www.ezbiocloud.net/app). (B) Comparison of C. acnes genomes. The RAST-annotated genomes of 6 C. acnes isolates (ATCC6919, HB, 2003-1719, 2004-10708, BL and Ntes) were aligned using C. acnes KPA171202 as a reference sequence. The genomes were aligned and the figure was created with BRIG software (http://brig.sourceforge.net/). Red arrows outside the outer circle indicate the locations of non-internalized strain-specific genes, blue arrows indicate the locations of internalized strain-specific genes.
Discussion
Most studies on the interaction between bacteria and bone cells during foreign-body infection have focused on the Staphylococcus aureus model, the main pathogen isolated in this context33,34,35. Very few other species have been studied and little is known about the impact of C. acnes either in aseptic loosening or low-grade infection36. In fact, C. acnes host-pathogen interactions have only been well demonstrated in the acne field, with a specific cutaneous innate immunity pattern according to phylotype37. To the best of our knowledge, this study is the first to provide information about the interaction and relationship between C. acnes and bone cells, in line with clinical and molecular typing data. Herein, using different lineages of C. acnes, we showed that strains belonging to the lineage previously associated with PJI (phylotype IB, CC36) were significantly less internalized by bone cells than those belonging to the lineage previously associated with acne or spine instrumentation infections (phylotype IA, CC18/CC28)13,17. We also showed that C. acnes can decrease the resorption ability of osteoclast with a greater effect of C. acnes belonging to CC36.
As shown in Fig. 5, both osteoblast and osteoclast interactions were studied to take into account the overall impact of this bacterium on the bone remodeling process. Osteoblasts, derived from mesenchymal bone marrow precursors, control this process by synthesizing the components of the bone matrix and by modulating the activity of the bone-resorbing osteoclasts1. Although bone cells can internalize bacteria by different mechanisms, i.e., phagocytosis for osteoclasts or endocytosis for osteoblasts38,39, interestingly, we observed a similar internalization behavior in response to C. acnes infection with osteoblasts, human mesenchymal stem cells (precursors of osteoblasts derived from bone marrow) and osteoclasts.
Depending on the clonal complexes, there are two ways for C. acnes to interact with bone cells. On one hand, CC18 C. acnes can be internalized by osteoblasts and osteoclasts. On the other hand, CC36 C. acnes are less internalized. Moreover, infection of mature osteoclasts by C. acnes decreases their bone resorption ability with a major impact by CC36 isolates. A question mark highlights the remaining issues to understand better the interaction between C. acnes and bone cells.
Using different epithelial cells, Mak et al. showed that a ST33/CC36 C. acnes strain (isolated from a prostatectomy tissue sample) was more internalized in RWPE1 cells than in HaCaT and HEKa cells40. Unfortunately, this could not be compared with our internalization level because only a relative invasion percentage was provided. On the other hand, two CC36 strains (P6 from cancerous prostate and KPA171202) and one CC18 strain (P266, pleuropulmonary isolate) showed strain-dependent differences in their uptake by the THP-1 cell line (macrophage model)41, without any correlation to the clade (45.9% and 8.9% of the initial inoculum for KPA and P6 strains, respectively). Taken together, these studies suggest that the CC36 strain trait observed is linked to the bone origin of the cells used in our study.
From a pathophysiological point of view, the evasion mechanism observed for CC36 C. acnes isolates could allow this clade to leave the site of infection, disseminate into deeper tissue layers and cause PJI (i.e., arthroplasty infection). Inside the deeper tissue, close to the material, the local immune defect fosters the low-grade infections observed42. In contrast, for CC18 and CC28 clades, mostly involved in spinal instrumentation infection, the internalization process observed could allow them to escape from the numerous immune cells located under the skin43 and generate an infection locally, favored by the spine instrumentation close to the skin. Moreover, the failure of osteoblasts to eradicate the intracellular CC18 C. acnes clade, resulting in bacterial persistence in the host, could explain the clinically observed BJI chronicity.
Additionally, we showed that direct infection of mature osteoclasts with C. acnes decreases their bone resorption ability. In vivo, a decrease in bone resorption will unbalance the bone remodeling process, leading to the possible aseptic loosening observed during C. acnes infection44. Interestingly, the CC36 C. acnes clade showed a major impact on osteoclast bone resorption ability. This result constitutes another argument to explain the deeper infection (PJI) tropism of CC36 C. acnes clades.
Finally, WGS analysis confirmed the high genome coverage and the highly stable chromosome previously observed45,46. All indels observed and linked to clade features (and most likely internalization) were recently described by Scholz et al.45. We identified 27 genes possibly linked to the behaviors observed (Table 2 in supplementary data). Perhaps the best candidate to explain our results is the hyaluronate lyase gene (indel_14)45. Indeed, the study of Tyner et al.47 and the recent communication from Nunes et al.48 highlighted that the allele diversity in the hyaluronate lyase gene could lead to different enzyme activities modulating, de facto, the penetration of C. acnes into tissues rich in hyaluronic acid (e.g. joints).
In conclusion, our study provides new data about C. acnes interactions with bone cells. Its strength is that it addresses the two main agents of the bone remodeling process with bacteria perfectly genetically defined due to the most recent molecular tools. We clearly identify two C. acnes behaviors depending on their genetic background (CC). These two behaviors provide explanations about the development of C. acnes PJI or spine instrumentation infection.
Additional Information
How to cite this article: Aubin, G. G. et al. Interaction of Cutibacterium (formerly Propionibacterium) acnes with bone cells: a step toward understanding bone and joint infection development. Sci. Rep. 7, 42918; doi: 10.1038/srep42918 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Deschaseaux, F., Sensébé, L. & Heymann, D. Mechanisms of bone repair and regeneration. Trends Mol Med 15, 417–429 (2009).
Beylot, C. et al. Propionibacterium acnes: an update on its role in the pathogenesis of acne. J Eur Acad Dermatol Venereol 28, 271–278 (2014).
Aubin, G. G., Portillo, M. E., Trampuz, A. & Corvec, S. Propionibacterium acnes, an emerging pathogen: from acne to implant-infections, from phylotype to resistance. Med Mal Infect 44, 241–250 (2014).
Achermann, Y., Goldstein, E. J. C., Coenye, T. & Shirtliff, M. E. Propionibacterium acnes: from Commensal to Opportunistic Biofilm-Associated Implant Pathogen. Clin. Microbiol. Rev. 27, 419–440 (2014).
Portillo, M. E., Corvec, S., Borens, O. & Trampuz, A. Propionibacterium acnes: An Underestimated Pathogen in Implant-Associated Infections. Biomed Res Int 2013, 804391 (2013).
Corvec, S., Portillo, M. E., Pasticci, B. M., Borens, O. & Trampuz, A. Epidemiology and new developments in the diagnosis of prosthetic joint infection. Int J Artif Organs 35, 923–934 (2012).
Osmon, D. R. et al. Diagnosis and management of prosthetic joint infection: clinical practice guidelines by the Infectious Diseases Society of America. Clin. Infect. Dis. 56, e1–e25 (2013).
Zappe, B., Graf, S., Ochsner, P. E., Zimmerli, W. & Sendi, P. Propionibacterium spp. in prosthetic joint infections: a diagnostic challenge. Arch Orthop Trauma Surg 128, 1039–1046 (2008).
Portillo, M. E. et al. Prosthesis failure within 2 years of implantation is highly predictive of infection. Clin. Orthop. Relat. Res. 471, 3672–3678 (2013).
Asseray, N. et al. Improving diagnostic criteria for Propionibacterium acnes osteomyelitis: a retrospective analysis. Scand. J. Infect. Dis. 42, 421–425 (2010).
Kilian, M., Scholz, C. F. P. & Lomholt, H. B. Multilocus sequence typing and phylogenetic analysis of Propionibacterium acnes . J. Clin. Microbiol. 50, 1158–1165 (2012).
McDowell, A. et al. An expanded multilocus sequence typing scheme for Propionibacterium acnes: investigation of ‘pathogenic’, ‘commensal’ and antibiotic resistant strains. PLoS ONE 7, e41480 (2012).
Lomholt, H. B. & Kilian, M. Population genetic analysis of Propionibacterium acnes identifies a subpopulation and epidemic clones associated with acne. PLoS ONE 5, e12277 (2010).
McDowell, A. et al. A novel multilocus sequence typing scheme for the opportunistic pathogen Propionibacterium acnes and characterization of type I cell surface-associated antigens. Microbiology (Reading, Engl.) 157, 1990–2003 (2011).
Scholz, C. F. P., Jensen, A., Lomholt, H. B., Brüggemann, H. & Kilian, M. A Novel High-Resolution Single Locus Sequence Typing Scheme for Mixed Populations of Propionibacterium acnes In Vivo . PLoS ONE 9, e104199 (2014).
Fitz-Gibbon, S. et al. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J. Invest. Dermatol. 133, 2152–2160 (2013).
Sampedro, M. F. et al. Species of Propionibacterium and Propionibacterium acnes phylotypes associated with orthopedic implants. Diagn. Microbiol. Infect. Dis. 64, 138–145 (2009).
Yu, Y., Champer, J., Garbán, H. & Kim, J. Typing of Propionibacterium acnes: a review of methods and comparative analysis. Br. J. Dermatol. doi: 10.1111/bjd.13667 (2015).
Bémer, P. et al. Significance of Propionibacterium acnes-positive samples in spinal instrumentation. Spine 33, E971–976 (2008).
Bémer, P. et al. Evaluation of 16S rRNA gene PCR sensitivity and specificity for diagnosis of prosthetic joint infection: a prospective multicenter cross-sectional study. J. Clin. Microbiol. 52, 3583–3589 (2014).
Osmon, D. R. et al. Diagnosis and management of prosthetic joint infection: clinical practice guidelines by the Infectious Diseases Society of America. Clin. Infect. Dis. 56, e1–e25 (2013).
Mollerup, S. et al. Propionibacterium acnes: Disease-Causing Agent or Common Contaminant? Detection in Diverse Patient Samples by Next-Generation Sequencing. J. Clin. Microbiol. 54, 980–987 (2016).
Darling, A. C. E., Mau, B., Blattner, F. R. & Perna, N. T. Mauve: Multiple Alignment of Conserved Genomic Sequence With Rearrangements. Genome Res. 14, 1394–1403 (2004).
Aziz, R. K. et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genomics 9, 75 (2008).
Alikhan, N. -F., Petty, N. K., Ben Zakour, N. L. & Beatson, S. A. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12, 402 (2011).
Lee, I., Kim, Y. O., Park, S.-C. & Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol, doi: 10.1099/ijsem.0.000760 (2015).
Guihard, P. et al. Induction of osteogenesis in mesenchymal stem cells by activated monocytes/macrophages depends on oncostatin M signaling. Stem Cells 30, 762–772 (2012).
Crémet, L. et al. Pathogenic potential of Escherichia coli clinical strains from orthopedic implant infections towards human osteoblastic cells. Pathog Dis 73, ftv065 (2015).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Duplomb, L. et al. Interleukin-6 inhibits receptor activator of nuclear factor kappaB ligand-induced osteoclastogenesis by diverting cells into the macrophage lineage: key role of Serine727 phosphorylation of signal transducer and activator of transcription 3. Endocrinology 149, 3688–3697 (2008).
Aubin, G. G. et al. Draft Genome Sequence of an Erythromycin-Resistant Propionibacterium acnes Isolate Recovered from Folliculitis of the Scalp. Genome Announc 5(4) (2017).
Aubin, G. G. et al. Draft Genome Sequences of Four Propionibacterium acnes Strains, Isolated from Implant-Related Infections. Genome Announc 4(6) (2016).
Trouillet-Assant, S. et al. Adaptive processes of Staphylococcus aureus isolates during the progression from acute to chronic bone and joint infections in patients. Cell. Microbiol, doi: 10.1111/cmi.12582 (2016).
Trouillet-Assant, S. et al. Dual impact of live Staphylococcus aureus on the osteoclast lineage, leading to increased bone resorption. J. Infect. Dis. 211, 571–581 (2015).
Aubin, G. G. et al. Staphylokinase and ABO group phenotype: new players in Staphylococcus aureus implant-associated infections development. Future Microbiol 10, 1929–1938 (2015).
Trouillet, S. et al. A novel flow cytometry-based assay for the quantification of Staphylococcus aureus adhesion to and invasion of eukaryotic cells. J. Microbiol. Methods 86, 145–149 (2011).
Jasson, F. et al. Different strains of Propionibacterium acnes modulate differently the cutaneous innate immunity. Exp. Dermatol. 22, 587–592 (2013).
Claro, T. et al. Staphylococcus aureus protein A binds to osteoblasts and triggers signals that weaken bone in osteomyelitis. PLoS ONE 6, e18748 (2011).
Heymann, D., Guicheux, J. & Rousselle, A. V. Ultrastructural evidence in vitro of osteoclast-induced degradation of calcium phosphate ceramic by simultaneous resorption and phagocytosis mechanisms. Histol. Histopathol. 16, 37–44 (2001).
Mak, T. N. et al. Propionibacterium acnes host cell tropism contributes to vimentin-mediated invasion and induction of inflammation. Cell. Microbiol. 14, 1720–1733 (2012).
Fischer, N. et al. Deciphering the intracellular fate of Propionibacterium acnes in macrophages. Biomed Res Int 2013, 603046 (2013).
Zimmerli, W., Lew, P. D. & Waldvogel, F. A. Pathogenesis of foreign body infection. Evidence for a local granulocyte defect. J. Clin. Invest. 73, 1191–1200 (1984).
Dréno, B. et al. Understanding innate immunity and inflammation in acne: implications for management. J Eur Acad Dermatol Venereol 29 Suppl 4, 3–11 (2015).
Feng, X. & McDonald, J. M. Disorders of Bone Remodeling. Annu Rev Pathol 6, 121–145 (2011).
Scholz, C. F. P., Brüggemann, H., Lomholt, H. B., Tettelin, H. & Kilian, M. Genome stability of Propionibacterium acnes: a comprehensive study of indels and homopolymeric tracts. Sci Rep 6, 20662 (2016).
Tomida, S. et al. Pan-genome and comparative genome analyses of Propionibacterium acnes reveal its genomic diversity in the healthy and diseased human skin microbiome. MBio 4, e00003–00013 (2013).
Tyner, H. & Patel, R. Hyaluronidase in Clinical Isolates of Propionibacterium acnes . Int J Bacteriol 2015 (2015).
Nunes, S. & Nazipi, S. The role of hyaluronate lyases of Propionibacterium acnes in tissue colonization and inflammation. ECCMID Amsterdam (2016).
Acknowledgements
We would like to thank: Stanimir Kambarev and Frédéric Pécorari (CRCINA, INSERM, CNRS, Université d’Angers, Université de Nantes, Nantes France for fruitful discussions and help, especially concerning WGS. Brigitte Dréno (Dermatology and Cancerology Unit, CHU NANTES) for providing acne lesion samples. Philippe Hulin and Steven Nedelec (Micropicell platform, Nantes University Hospital) for technical assistance and helpful discussions regarding confocal microscopy and video imaging. The CRIOGO study group (Cécile Lebrun (Tours University Hospital), Didier Tandé (Brest University Hospital), Chloé Pouzeau (Poitiers University Hospital), Anne Jolivet-Gougeon (Rennes University Hospital)) and Sandra Bourdon (La Roche/Yon Hospital) for providing C. acnes clinical isolates. Audrey Renaud and Céline Charrier (INSERM, UMR 957, Nantes University) for technical assistance. The Geno BiRD core facility for its technical support.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: S.C. and D.H. Performed the experiments: G.A., M.B. and R.B. Analyzed the data: G.A., S.C., J.P.L. and M.B. Contributed reagents/materials/analysis tools: S.C., F.G., K.A. and D.H. Wrote the paper: G.A. Edited the paper: S.C., J.P.L., C.J., K.A., D.L., M.B. and D.H. Statistical analysis: G.A.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Aubin, G., Baud’huin, M., Lavigne, JP. et al. Interaction of Cutibacterium ( formerly Propionibacterium) acnes with bone cells: a step toward understanding bone and joint infection development. Sci Rep 7, 42918 (2017). https://doi.org/10.1038/srep42918
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep42918
This article is cited by
-
Intervertebral disc puncture predisposes the disc to Cutibacterium acnes infection via hematogenous and contiguous routes: a rat model study
Journal of Orthopaedic Surgery and Research (2026)
-
Cutibacterium acnes biofilm formation is influenced by bone microenvironment, implant surfaces and bacterial internalization
BMC Microbiology (2024)
-
Structural stability of Cutibacterium acnes acyl carrier protein studied using CD and NMR spectroscopy
Journal of Analytical Science and Technology (2022)
-
Misidentification of Cutibacterium namnetense as Cutibacterium acnes among clinical isolates by MALDI-TOF VitekMS: usefulness of gyrB sequencing and new player in bone infections
European Journal of Clinical Microbiology & Infectious Diseases (2020)
-
Titanium granules pre-treated with hydrogen peroxide inhibit growth of bacteria associated with post-operative infections in spine surgery
European Spine Journal (2018)