Abstract
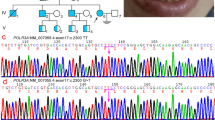
Apolipoprotein B 3′ (3′ ApoB) minisatellite polymorphism was studied in healthy unrelated individuals from the Russian Federation and the Republic of Belarus, in 10 populations from five ethnic groups: Russians, Byelorussians, Adygeis, Kalmyks and Yakuts. The analysis was carried out using PCR and electrophoresis followed by silver staining. Overall, 25 alleles of the 3′ ApoB minisatellite, ranging from 25 to 55 repeats, were detected. Heterozygosity indices were high and varied from 0.73 to 0.84. The distributions of alleles of this minisatellite in the Caucasoid populations (Russians, Byelorussians and Adygeis) had a bimodal character, whereas that for Mongoloid populations (Kalmyks and Yakuts) had a unimodal distribution. Nei's genetic distances between the populations studied and some reference populations of Europe and Asia were estimated. Despite their allele distribution homogeneity, different East Slavonic ethnic groups were clearly resolved by multidimensional analyses. The East Slavonic and Adygei populations revealed a high similarity with European Caucasoids. The Mongoloid populations (Kalmyks and Yakuts) were considerably different from those of the European Caucasoid populations, but were similar to other Asian Mongoloid populations. The results demonstrate the variability of 3′ ApoB minisatellite polymorphism not only in distant populations but also, to a certain extent, in genetically relative ones.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Jeffreys AJ, Wilson V, Thein SL: Hypervariable minisatellite regions in human DNA. Nature 1985; 314: 67–73.
Destro-Bisol G, Capelli C, Belledi M: Inferring microevolutionary patterns from allele-size frequency distributions of minisatellite loci: a worldwide study of the APOB 3′ hypervariable region polymorphism. Hum Biol 2000; 72: 733–751.
Chen B, Guo Z, He P, Ye P, Buresi C, Roizes G: Structure and function of alleles in the 3′ end region of human apoB gene. Chin Med J (Engl) 1999; 112: 221–223.
Huang LS, Breslow JL: A unique AT-rich hypervariable minisatellite 3′ to the ApoB gene defines a high information restriction fragment length polymorphism. J Biol Chem 1987; 262: 8952–8955.
Buresi C, Desmarais E, Vigneron S et al: Structural analysis of the minisatellite present at the 3′ end of the human apolipoprotein B gene: new definition of the alleles and evolutionary implications. Hum Mol Genet 1996; 5: 61–68.
Marz W, Ruzicka V, Fisher E, Russ AP, Schnider W, Gross W: Typing of the 3′ hypervariable region of the apolipoprotein B gene: approaches, pitfalls and applications. Electrophoresis 1993; 14: 169–173.
Renges HH, Peacock R, Dunning AM, Talmud P, Humphries SE: Genetic relationship between the 3′ VNTR and diallelic apolipoprotein B gene polymorphism: haplotype analysis in individuals of European and South Asian origin. Ann Hum Genet 1992; 56: 11–33.
Kravchenko SA, Malyarchuk SG, Livshits LA: A population genetics study of the allelic polymorphism in the hypervariable region of the apolipoprotein B gene in the population of different regions of Ukraine. Tsitology Genet 1996; 30: 35–41.
De Benedictis G, Carotenuto L, Carrieri G et al: Age-related changes of the 3′APOB-VNTR genotype pool in ageing cohorts. Ann Hum Genet 1998; 62: 115–122.
Kadasi L, Gecz J, Ferakova I et al: Distribution of ApoBII, MCT118 (D1S80), YNZ22 (D17S30) and COL2A1 Amp-FLPs (amplified fragment length polymorphisms) in Caucasoid population of Slovakia. Gene Geography 1994; 8: 121–127.
Friedl W, Ludwig EH, Paulweber B, Sandhofer F, McCarthy BJ: Hypervariability in a minisatellite 3′ of the apolipoprotein B gene in patients with coronary heart disease compared with normal control. J Lipid Res 1990; 31: 659–665.
Poltl R, Luckenbach C, Reinhold J, Fimmers R, Ritter H: Comparison of German population data on the apoB-HVR locus with other Caucasian, Asian and Black populations. Forensic Sci Int 1996; 80: 221–227.
Domenici R, Fornaciari S, Nardone M et al: Study of the ApoB polymorphism in Tuscany (Italy); in Bar W, Fiori A, Rossi U (eds): Advances in forensic haemogenetics. Berlin: Springer, 1995, pp 493–495.
Alonso S, Fernandez I, Castro A et al: Genetic characterization of APOB and D17S5 AFLP loci in a sample from the Basque Country (northern Spain). Hum Biol 1998; 70: 491–505.
Pestoni C, Munoz I, Comesana M et al: Distribution of the AmpFLPs YNZ22, 3′ApoB and COL2A1 in the population of Galicia (NW Spain). Forensic Sci Int 1996; 80: 175–188.
Pinheiro MF, Pontes ML, Huguet E, Gene M, da Costa JP, Moreno P: Study of three AMPFLPs (D1S80, 3′ApoB and YNZ22) in the population of the north of Portugal. Forensic Sci Int 1996; 79: 23–29.
Lahermo P, Sajantila A, Sistonen P et al: The genetic relationship between the Finns and the Finnish Saami (Lapps): analysis of nuclear DNA and mtDNA. Am J Hum Genet 1996; 58: 1309–1322.
Woller J, Furedi S, Padar Z: AMPFLP analysis of the VNTR loci and ApoB in Hungary. Int J Leg Med 1995; 107: 273–274
Zago MA, Silva Jr WA, Tavella MH, Santos SE, Guerreiro JF, Figueiredo MS: Interpopulational genetic diversity of Amerindians as revealed by six variable number of tandem repeats. Hum Hered 1996; 46: 274–289.
Evans AE, Zhang W, Moreel JFR et al: Polymorphisms of the apolipoprotein B and E genes and their relationship to plasma lipid variables in healthy Chinese man. Hum Genet 1993; 92: 191–197.
Alekseev VP: The morphological specificity of Caucasian peoples according to craniological materials. J Hum Evol 1982; 11: 663–672.
Milligan BG: Total DNA isolation. in: Hoelzel AR (ed). Molecular genetics analysis of populations. London: Oxford University Press, 1998, pp 29–60.
Schlotterer C: Minisatellites. In: Hoelzel AR (ed). Molecular genetics analysis of populations. London: Oxford University Press, 1998. pp 237–260.
Sajantila A, Puomilahti S, Johnsson V, Ehnholm C: Amplification of reproducible allele markers for amplified fragment length polymorphism analysis. Biotechniques 1992; 12: 16–20.
Pogoda TV, Nikonova AL, Kolosova TV, Liudvikova EK, Perova NV, Limborska SA: Allelic variants of apolipoproteins B and CII genes in patients with ischemic heart disease and in healthy persons from the Moscow population. Russ J Genet 1995; 31: 1001–1009.
Ludwig EH, Friedl W, McCarthy BJ: High resolution analysis of a hypervariable region in the human apolipoprotein B gene. Am J Hum Genet 1989; 45: 458–464.
Yeh FC, Yang R-C, Boyle T: POPGENE version 1.32. 1999. http://www.ualberta.ca/~fyeh/
Sokal RR, Rohlf JF: Biometry, 2nd ed. New York: Freeman, 1969.
Guo SW, Thompson EA . Performing the exact test of Hardy–Weinberg proportion for multiple alleles. Biometrics 1992; 48: 361–372.
Roff DA, Bentzen P: The statistical analysis of mitochondrial DNA: χ2 and problem of small samples. Mol Biol Evol 1989; 6: 539–545.
STATISTICA for windows (computer program manual). Tulsa, OK: StatSoft, Inc., 2000. http://www.statsoft.com
Weir BS: Genetic data analysis. Sunderland, MA: Sinauer Associates, 1990.
Nei M: Genetic distance between populations. Am Nat 1972; 106: 283–292.
Sedov VV: Slavs in antiquity. Moscow: Archaeological Fund, 1994.
Alekseeva TI, Alekseev VP: Ethnogeny of Slavic peoples: an anthropologist's view. Ethnologia Slavica (Bratislava) 1977; 8–9: 13–23.
Malyarchuk BA, Derenko MV: Mitochondrial DNA variability in Russians and Ukrainians: implication to the origin of the Eastern Slavs. Ann Human Genet 2001; 65: 63–78.
Kravchenko SA, Slominsky PA, Bets LA et al: Polymorphism of STR loci of the Y chromosome in three populations of Eastern Slavs from Belarus, Russia and Ukraine. Russ J Genet 2002; 38: 97–104.
Laitinen V, Lahermo P, Sistonen P, Savontaus M-L: Chromosomal diversity suggests that Baltic males share common Finno-Ugric-speaking forefathers. Hum Heredity 2002; 53: 68–78.
Macaulay V, Richards M, Hickey E et al: The emerging tree of West Eurasian mtDNAs: a synthesis of control-region sequences and RFLPs. Am J Hum Genet 1999; 64: 232–249.
Cavalli-Sforza LL, Menozzi P, Piazza A: The history and geography of human genes. Princeton, NJ: Princeton University Press, 1994.
Galushkin SK, Spitsyn VA, Crawford MH: Genetic structure of Mongolic-speaking Kalmyks. Hum Biol 2001; 73: 823–834.
Acknowledgements
We thank Professor Giovanni Destro-Bisol who contributed the 3′ ApoB allele frequencies of numerous populations. We are indebted to Professor Vasily Deryabin for valuable comments and discussion. We are also grateful to two anonymous reviewers whose comments and suggestions significantly improved the manuscript. This study was supported by grants from the Russian Basic Research Foundation, and the Russian Foundation for Humanities.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Verbenko, D., Pogoda, T., Spitsyn, V. et al. Apolipoprotein B 3′-VNTR polymorphism in Eastern European populations. Eur J Hum Genet 11, 444–451 (2003). https://doi.org/10.1038/sj.ejhg.5200986
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/sj.ejhg.5200986
Keywords
This article is cited by
-
Genetic association of APOB polymorphisms with variation in serum lipid profile among the Kuwait population
Lipids in Health and Disease (2014)
-
Effect of the 3'APOB-VNTR polymorphism on the lipid profiles in the Guangxi Hei Yi Zhuang and Han populations
BMC Medical Genetics (2007)
-
p53 polymorphisms in Russia and Belarus: correlation of the 2-1-1 haplotype frequency with longitude
Molecular Genetics and Genomics (2005)