Abstract
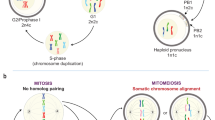
Somatic cell nuclear transfer (SCNT) and parthenogenesis are alternative forms of reproduction and development, building new life cycles on differentiated somatic cell nuclei and duplicated maternal chromatin, respectively. In the preceding paper (Sun F, et al., Cell Res 2007; 17:117–134.), we showed that an “erase-and-rebuild” strategy is used in normal development to transform the maternal gene expression profile to a zygotic one. Here, we investigate if the same strategy also applies to SCNT and parthenogenesis. The relationship between chromatin and chromatin factors (CFs) during SCNT and parthenogenesis was examined using immunochemical and GFP-fusion protein assays. Results from these studies indicated that soon after nuclear transfer, a majority of CFs dissociated from somatic nuclei and were redistributed to the cytoplasm of the egg. The erasure process in oogenesis is recaptured during the initial phase in SCNT. Most CFs entered pseudo-pronuclei shortly after their formation. In parthenogenesis, all parthenogenotes underwent normal oogenesis, and thus had removed most CFs from chromosomes before the initiation of development. The CFs were subsequently re-associated with female pronuclei in time and sequence similar to that in fertilized embryos. Based on these data, we conclude that the “erase-and-rebuild” process observed in normal development also occurs in SCNT and in parthenogenesis, albeit in altered fashions. The process is responsible for transcription reprogramming in these procedures. The “erase” process in SCNT is compressed and the efficiency is compromised, which likely contribute to the developmental defects often observed in nuclear transfer (nt) embryos. Furthermore, results from this study indicated that the cytoplasm of an egg contains most, if not all, essential components for assembling the zygotic program and can assemble them onto appropriate diploid chromatin of distinct origins.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Meissner A, Jaenisch R . Mammalian nuclear transfer. Dev Dyn 2006; 235:2460–2469.
Collas P, Pinto-Correia C, Ponce de Leon FA, Robl JM . Effect of donor cell cycle stage on chromatin and spindle morphology in nuclear transplant rabbit embryos. Biol Reprod 1992; 46:501–511.
Campbell KH, Ritchie WA, Wilmut I . Nuclear-cytoplasmic interactions during the first cell cycle of nuclear transfer reconstructed bovine embryos: implications for deoxyribonucleic acid replication and development. Biol Reprod 1993; 49:933–942.
Gurdon JB, Laskey RA, De Robertis EM, Partington GA . Reprogramming of transplanted nuclei in amphibia. Int Rev Cytol Suppl 1979:161–178.
Eggan K, Akutsu H, Hochedlinger K, Rideout W III, Yanagimachi R, Jaenisch R . X-Chromosome inactivation in cloned mouse embryos. Science 2000; 290:1578–1581.
Lanza RP, Cibelli JB, Blackwell C, et al. Extension of cell life-span and telomere length in animals cloned from senescent somatic cells. Science 2000; 288:665–669.
Dean W, Santos F, Stojkovic M, et al. Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos. Proc Natl Acad Sci USA 2001; 98:13734–13738.
Kang YK, Koo DB, Park JS, et al. Aberrant methylation of donor genome in cloned bovine embryos. Nat Genet 2001; 28:173–177.
Humpherys D, Eggan K, Akutsu H, et al. Epigenetic instability in ES cells and cloned mice. Science 2001; 293:95–97.
Bourc'his D, Le Bourhis D, Patin D, et al. Delayed and incomplete reprogramming of chromosome methylation patterns in bovine cloned embryos. Curr Biol 2001; 11:1542–1546.
Inoue K, Kohda T, Lee J, et al. Faithful expression of imprinted genes in cloned mice. Science 2002; 295:297.
Kang YK, Park JS, Koo DB, et al. Limited demethylation leaves mosaic-type methylation states in cloned bovine pre-implantation embryos. EMBO J 2002; 21:1092–1100.
Gurdon JB . Nuclear transplantation in eggs and oocytes. J Cell Sci Suppl 1986; 4:287–318.
Kim JM, Ogura A, Nagata M, Aoki F . Analysis of the mechanism for chromatin remodeling in embryos reconstructed by somatic nuclear transfer. Biol Reprod 2002; 67:760–766.
Ng RK, Gurdon JB . Epigenetic memory of active gene transcription is inherited through somatic cell nuclear transfer. Proc Natl Acad Sci USA 2005; 102:1957–1962.
Surani MA . Nuclear reprogramming by human embryonic stem cells. Cell 2005; 122:653–654.
Hochedlinger K, Jaenisch R . Nuclear reprogramming and pluripotency. Nature 2006;441:1061–1067.
Gurdon JB, Byrne JA . The first half-century of nuclear transplantation. Biosci Rep 2004; 24:545–557.
Vrana KE, Hipp JD, Goss AM, et al. Nonhuman primate parthenogenetic stem cells. Proc Natl Acad Sci USA 2003; 100 Suppl 1:11911–11916.
Surani MA, Barton SC . Development of gynogenetic eggs in the mouse: implications for parthenogenetic embryos. Science 1983; 222:1034–1036.
Surani MA, Kothary R, Allen ND, et al. Genome imprinting and development in the mouse. Dev Suppl 1990; 89–98.
McGrath J, Solter D . Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 1984; 37:179–183.
Kure-bayashi S, Miyake M, Okada K, Kato S . Successful implantation of in vitro-matured, electro-activated oocytes in the pig. Theriogenology 2000; 53:1105–1119.
Hagemann LJ, Peterson AJ, Weilert LL, Lee RS, Tervit HR . In vitro and early in vivo development of sheep gynogenones and putative androgenones. Mol Reprod Dev 1998; 50:154–162.
Barton SC, Surani MA, Norris ML . Role of paternal and maternal genomes in mouse development. Nature 1984; 311:374–376.
Kono T . Genomic imprinting is a barrier to parthenogenesis in mammals. Cytogenet Genome Res 2006; 113:31–35.
Kono T, Obata Y, Wu Q, et al. Birth of parthenogenetic mice that can develop to adulthood. Nature 2004; 428:860–864.
Chatot CL, Ziomek CA, Bavister BD, Lewis JL, Torres I . An improved culture medium supports development of random-bred 1-cell mouse embryos in vitro. J Reprod Fertil 1989; 86:679–688.
Adenot PG, Mercier Y, Renard JP, Thompson EM . Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development 1997; 124:4615–4625.
Wakayama T, Perry AC, Zuccotti M, Johnson KR, Yanagimachi R . Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature 1998; 394:369–374.
Inoue K, Ogonuki N, Mochida K, et al. Effects of donor cell type and genotype on the efficiency of mouse somatic cell cloning. Biol Reprod 2003; 69:1394–1400.
Worrad DM, Ram PT, Schultz RM . Regulation of gene expression in the mouse oocyte and early preimplantation embryo: developmental changes in Sp1 and TATA box-binding protein, TBP. Development 1994; 120:2347–2357.
Wang K, Sun F, Sheng HZ . Regulated expression of TAF1 in 1-cell mouse embryos. Zygote 2006; 14:209–215.
Sun F, Tang F, Yan AY, Fang HY, Sheng HZ . Expression of SRG3, a chromatin remodeling factor, in the mouse oocyte and early preimplantation embryos. Zygote 2007; 15:1–10.
Kikyo N, Wade PA, Guschin D, Ge H, Wolffe AP . Active remodeling of somatic nuclei in egg cytoplasm by the nucleosomal ATPase ISWI. Science 2000; 289:2360–2362.
Ecker RE, Smith LD . The nature and fate of Rana pipiens proteins synthesized during maturation and early cleavage. Dev Biol 1971; 24:559–576.
Gurdon JB, De Robertis EM, Partington G . Injected nuclei in frog oocytes provide a living cell system for the study of transcriptional control. Nature 1976; 260:116–120.
Simonsson S, Gurdon J . DNA demethylation is necessary for the epigenetic reprogramming of somatic cell nuclei. Nat Cell Biol 2004; 6:984–990.
Kim JM, Liu H, Tazaki M, Nagata M, Aoki F . Changes in histone acetylation during mouse oocyte meiosis. J Cell Biol 2003; 162:37–46.
Lemaitre JM, Danis E, Pasero P, Vassetzky Y, Mechali M . Mitotic remodeling of the replicon and chromosome structure. Cell 2005; 123:787–801.
Sun F, Fang H, Li R, et al. Nuclear reprogramming: the zygotic transcription program is established through an “erase-and-rebuild” strategy. Cell Res 2007; Feb 6. [Epub ahead of print]
Acknowledgements
We are grateful to Dr Yun-Bo Shi, Yingzi Yang, and Paul Zhou for criticizing the manuscript. We are thankful to Dr Shangang Li for assistance in nuclear transfer procedure, Wei Su for artwork, Youming Zhu for cell culture, Wei Liu for assistance in immunochemistry, and Fengying Li, Wenqin Ying, and Wanli Li for animal care. We also apologize for citing reviews instead of original publications in some places due to space limitations. Please look in the reviews for original papers.
The study was supported by grants from National Basic Research Program of China (973 Program) (No. 001CB509903, 001CB509904), Hi-Tech Research and Development Program of China (863 Program) (No. 2001AA216121, 2004AA205010), Science and Technology Committee of Shanghai Municipality (No. 99DJ14002, 00DJ1 4033, 01DJ14003, 03DJ14017), Shanghai Municipal Education Commission (No. T0205) and Shanghai Jiao Tong University, School of Medicine.
Contributions: Tianlong Gao and Haiyan Fang are responsible for GFP-CF analyses, Junke Zheng, Feng Sun, Ayong Yan, Xun Gong, Hui Ding, and Fang Tang for analyses on nt-embryos, Fengying Xing for analyses in parthenogenotes. Feng Sun assisted in manuscript writing. Hui Z Sheng is responsible for development of the model, project planning, and most manuscript writing.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gao, T., Zheng, J., Xing, F. et al. Nuclear reprogramming: the strategy used in normal development is also used in somatic cell nuclear transfer and parthenogenesis. Cell Res 17, 135–150 (2007). https://doi.org/10.1038/cr.2007.2
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/cr.2007.2
Keywords
This article is cited by
-
KDM4B-mediated reduction of H3K9me3 and H3K36me3 levels improves somatic cell reprogramming into pluripotency
Scientific Reports (2017)
-
Epigenetic modification with trichostatin A does not correct specific errors of somatic cell nuclear transfer at the transcriptomic level; highlighting the non-random nature of oocyte-mediated reprogramming errors
BMC Genomics (2016)
-
Nuclear reprogramming of sperm and somatic nuclei in eggs and oocytes
Reproductive Medicine and Biology (2013)
-
Mechanisms of nuclear reprogramming by eggs and oocytes: a deterministic process?
Nature Reviews Molecular Cell Biology (2011)
-
Nuclear reprogramming: the zygotic transcription program is established through an “erase-and-rebuild” strategy
Cell Research (2007)