Abstract
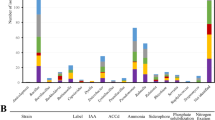
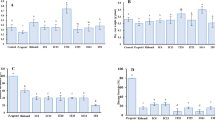
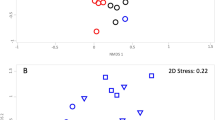
We constructed an experimental model system to study the effects of grazing by a common soil amoeba, Acanthamoeba castellanii, on the composition of bacterial communities in the rhizosphere of Arabidopsis thaliana. Amoebae showed distinct grazing preferences for specific bacterial taxa, which were rapidly replaced by grazing tolerant taxa in a highly reproducible way. The relative proportion of active bacteria increased although bacterial abundance was strongly decreased by amoebae. Specific bacterial taxa had disappeared already two days after inoculation of amoebae. The decrease in numbers was most pronounced in Betaproteobacteria and Firmicutes. In contrast, Actinobacteria, Nitrospira, Verrucomicrobia and Planctomycetes increased. Although other groups, such as betaproteobacterial ammonia oxidizers and Gammaproteobacteria did not change in abundance, denaturing gradient gel electrophoresis with specific primers for pseudomonads (Gammaproteobacteria) revealed both specific changes in community composition as well as shifts in functional genes (gacA) involved in bacterial defence responses. The resulting positive feedback on plant growth in the amoeba treatment confirms that bacterial grazers play a dominant role in structuring bacteria–plant interactions. This is the first detailed study documenting how rapidly protozoan grazers induce shifts in rhizosphere bacterial community composition.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Acea MJ, Moore CR, Alexander M . (1988). Survival and growth of bacteria introduced into soil. Soil Biol Biochem 20: 509–515.
Alphei J, Bonkowski M, Scheu S . (1996). Protozoa, Nematoda and Lumbricidae in the rhizosphere of Hordelymus Europaeus (Poaceae): faunal interactions, response of microorganisms and effects on plant growth. Oecologia 106: 111–126.
Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA . (1990). Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol 56: 1919–1925.
Andersen KS, Winding A . (2004). Non-target effects of bacterial biological control agents on soil protozoa. Biol Fertil Soils 40: 230–236.
Bertaux J, Gloger U, Schmidt M, Hartmann A, Scheu S . (2007). Routine fluorescence in situ hybridization in soil. J Microbiol Methods 69: 451–460.
Bjørnlund L, Mork S, Vestergard M, Rønn R . (2006). Trophic interactions between rhizosphere bacteria and bacterial feeders influenced by phosphate and aphids in barley. Biol Fertil Soils 43: 1–11.
Bonkowski M . (2004). Protozoa and plant growth: the microbial loop in soil revisited. New Phytol 162: 617–631.
Bonkowski M, Brandt F . (2002). Do soil protozoa enhance plant growth by hormonal effects? Soil Biol Biochem 34: 1709–1715.
Bonkowski M, Cheng WX, Griffiths BS, Alphei J, Scheu S . (2000). Microbial-faunal interactions in the rhizosphere and effects on plant growth. Eur J Soil Biol 36: 135–147.
Brosius J, Dull TL, Sleeter DD, Noller HF . (1981). Gene organization of primary structure of a ribosomal operon from Escherichia coli. J Mol Biol 148: 107–127.
Clarholm M . (1981). Protozoan grazing of bacteria in soil—impact and importance. Microb Ecol 7: 343–350.
Clarholm M . (1989). Effects of plant-bacterial-amoebal interactions on plant uptake of nitrogen under field conditions. Biol Fertil Soils 8: 373–378.
Costa R, Salles JF, Berg G, Smalla K . (2006). Cultivation-independent analysis of Pseudomonas species in soil and in the rhizosphere of field-grown Verticillium dahliae host plants. Environ Microbiol 8: 2136–2149.
Daims H, Brühl A, Amann R, Schleifer KH, Wagner M . (1999). Probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol 22: 438–448.
Daims H, Nielsen JL, Nielsen PH, Schleifer K-H, Wagner M . (2001). In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl Environ Microbiol 67: 5273–5284.
Danso SKA, Alexander M . (1975). Regulation of predation by prey density-protozoan-rhizobium relationship. Appl Microbiol 29: 515–521.
Darbyshire JF, Wheatley RE, Greaves MP, Inkson RHE . (1974). Rapid micromethod for estimating bacterial and protozoan populations in soil. Rev Ecol Biol Sol 11: 465–475.
De Leij FAAM, Dixon-Hardy JE, Lynch JM . (2002). Effect of 2,4-diacetylphloroglucinol-producing and non-producing strains of Pseudomonas fluorescens on root development of pea seedlings in three different soil types and its effect on nodulation by Rhizobium. Biol Fertil Soils 35: 114–121.
De Souza JT, Mazzola M, Raaijmakers JM . (2003). Conservation of the response regulator gene gacA in Pseudomonas species. Environ Microbiol 5: 1328–1340.
Finlay BF, Black HIJ, Brown S, Clarke KJ, Esteban GF, Hindle RM et al. (2000). Estimating the growth potential of the soil protozoan community. Protist 151: 69–80; Erratum Protist 151: 367.
Foster RC, Dormaar JF . (1991). Bacteria-grazing amoebas in situ in the rhizosphere. Biol Fertil Soils 11: 83–87.
Griffiths BS . (1989). Enhanced nitrification in the presence of bacteriophagous protozoa. Soil Biol Biochem 21: 1045–1051.
Griffiths BS, Bonkowski M, Dobson G, Caul S . (1999). Changes in soil microbial community structure in the presence of microbial-feeding nematodes and protozoa. Pedobiologia 43: 297–304.
Habte M, Alexander M . (1975). Protozoa as agents responsible for decline of Xanthomonas campestris in soil. Appl Microbiol 29: 159–164.
Hahn MW, Höfle MG . (1998). Grazing pressure by a bacterivorous flagellate reverses the relative abundance of Comamonas acidovorans Px54 and Vibrio Strain Cb5 in chemostat co-cultures. Appl Environ Microbiol 64: 1910–1918.
Hurley MA, Roscoe ME . (1983). Automated statistical-analysis of microbial enumeration by dilution series. J Appl Bacteriol 55: 159–164.
Janssen PH . (2006). Identifying the dominant soil bacterial taxa in libraries of 16 s rRNA and 16 s rRNA genes. Appl Environ Microbiol 72: 1719–1728.
Jossi M, Fromin N, Tarnawski S, Kohler F, Gillet F, Aragno M et al. (2006). How elevated pCO2 modifies total and metabolically active bacterial communities in the rhizosphere of two perennial grasses grown under field conditions. FEMS Microbiol Ecol 55: 339–350.
Jousset A, Lara E, Wall LG, Valverde C . (2006). Secondary metabolites help biocontrol strain Pseudomonas fluorescens CHA0 to escape protozoan grazing. Appl Environ Microbiol 72: 7083–7090.
Jousset A, Scheu S, Bonkowski M . (2008). Secondary metabolite production facilitates establishment of rhizobacteria by reducing both protozoan predation and the competitive effects of indigenous bacteria. Funct Ecol 22: 714–719.
Jürgens K, Matz C . (2002). Predation as a shaping force for the phenotypic and genotypic composition of planktonic bacteria. Antonie Van Leeuwenhoek 81: 413–434.
Kreuzer K, Adamczyk J, Iijima M, Wagner M, Scheu S, Bonkowski M . (2006). Grazing of a common species of soil protozoa (Acanthamoeba castellanii) affects rhizosphere bacterial community composition and root architecture of rice (Oryza sativa L). Soil Biol Biochem 38: 1665–1672.
Krome K, Rosenberg K, Bonkowski M, Scheu S . (2009). Grazing of protozoa on rhizosphere bacteria alters growth and reproduction of Arabidopsis thaliana. New Phtytol (in press).
Lueders T, Manefield M, Friedrich MW . (2004). Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ Microbiol 6: 73–78.
Lugtenberg BJJ, Chin-a-Woeng TFC, Bloemberg GV . (2002). Microbe-plant interactions: principles and mechanisms. Antonie Van Leeuwenhoek 81: 373–383.
Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H . (1992). Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst Appl Microbiol 15: 593–600.
Mao XF, Hu F, Griffiths B, Chen XY, Liu MQ, Li HX . (2007). Do bacterial-feeding nematodes stimulate root proliferation through hormonal effects? Soil Biol Biochem 39: 1816–1819.
Mao XF, Hu F, Griffiths B, Li H . (2006). Bacterial-feeding nematodes enhance root growth of tomato seedlings. Soil Biol Biochem 38: 1615–1622.
Matz C, Kjelleberg S . (2005). Off the hook—how bacteria survive protozoan grazing. Trends Microbiol 13: 302–307.
Meier H, Amann R, Ludwig W, Schleifer KH . (1999). Specific oligonucleotide probes for in situ detection of a major group of Gram-positive bacteria with low DNA G+C content. Syst Appl Microbiol 22: 186–196.
Milling A, Smalla K, Maidl FX, Schloter M, Munch JC . (2004). Effects of transgenic potatoes with an altered starch composition on the diversity of soil and rhizosphere bacteria and fungi. Plant Soil 266: 23–39.
Mobarry BK, Wagner M, Urbain V, Rittmann BE, Stahl DA . (1996). Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl Environ Microbiol 62: 2156–2162. Erratum Appl Environ Microbiol 63: 815.
Moran NA, Russell JA, Koga R, Fukatsu T . (2005). Evolutionary relationships of three new species of Enterobacteriaceae living as symbionts of aphids and other insects. Appl Environ Microbiol 71: 3302–3310.
Murase J, Noll M, Frenzel P . (2006). Impact of protists on the activity and structure of the bacterial community in a rice field soil. Appl Environ Microbiol 72: 5436–5444.
O'Sullivan LA, Weightman AJ, Fry JC . (2002). New degenerate Cytophaga-Flexibacter-Bacteroides-specific 16S ribosomal DNA-targeted oligonucleotide probes reveal high bacterial diversity in River Taff epilithon. Appl Environ Microbiol 68: 201–210.
Page FC . (1976). An illustrated key to freshwater and soil amoebae. Freshwater Biological Assoc 34: 11.
Pernthaler J . (2005). Predation on prokaryotes in the water column and its ecological implications. Nat Rev Microbiol 3: 537–546.
Posch T, Simek K, Vrba J, Pernthaler S, Nedoma J, Sattler B et al. (1999). Predator-induced changes of bacterial size-structure and productivity studied on an experimental microbial community. Aquat Microb Ecol 18: 235–246.
Roller C, Wagner M, Amann R, Ludwig W, Schleifer K-H . (1994). In situ probing of Gram-positive bacteria with high DNA G+C content using 23S rRNA-targeted oligonucleotides. Microbiology 140: 2849–2858.
Rønn R, McCaig AE, Griffiths BS, Prosser JI . (2002). Impact of protozoan grazing on bacterial community structure in soil microcosms. Appl Environ Microbiol 68: 6094–6105.
Schuster FL . (2002). Cultivation of pathogenic and opportunistic free-living amoebas. Clin Microbiol Rev 15: 342–354.
Verhagen FJM, Duyts H, Laanbroek HJ . (1993). Effects of grazing by flagellates on competition for ammonium between nitrifying and heterotrophic bacteria in soil columns. Appl Environ Microbiol 59: 2099–2106.
Zhang HM, Forde BG . (1998). An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279: 407–409.
Zul D, Denzel S, Kotz A, Overmann J . (2007). Effects on plant biomass, plant diversity, and water content on bacterial communities in soil lysimeters: implications for the determinants of bacterial diversity. Appl Environ Microbiol 73: 6916–6929.
Acknowledgements
This study is part of the project ‘Virtual Institute of Biotic Interactions’ of the Helmholtz Association. We thank Dr Michael Schmidt, Helmholtz Zentrum München, Germany, for technical help with the clone libraries and Prof Dr Kornelia Smalla at Biologische Bundesanstalt (BBA) in Braunschweig, Germany, for her support in providing information on the gacA-specific primers.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rosenberg, K., Bertaux, J., Krome, K. et al. Soil amoebae rapidly change bacterial community composition in the rhizosphere of Arabidopsis thaliana. ISME J 3, 675–684 (2009). https://doi.org/10.1038/ismej.2009.11
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2009.11
Keywords
This article is cited by
-
Phagotrophic protists can change microbial nitrogen conversion patterns during swine manure composting
Biomass Conversion and Biorefinery (2024)
-
Contrasting response of rice rhizosphere microbiomes to in situ cadmium-contaminated soil remediation
Soil Ecology Letters (2024)
-
The Arabidopsis holobiont: a (re)source of insights to understand the amazing world of plant–microbe interactions
Environmental Microbiome (2023)
-
Long-term mowing reinforces connections between soil microbial and plant communities in a temperate steppe
Plant and Soil (2023)
-
Soil texture is a stronger driver of the maize rhizosphere microbiome and extracellular enzyme activities than soil depth or the presence of root hairs
Plant and Soil (2022)