Abstract
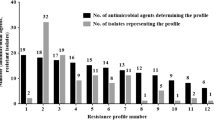
The quinolone resistance of 19 lactic acid bacterial strains belonging to the genera Enterococcus and Lactobacillus isolated from the natural fermented koumiss and yoghurt were investigated. The objective of this study was to determine the quinolone resistance levels and to explore the association of the resistance with the mutation patterns in gyrA and parC genes, as is currently recommended by the Food and Agriculture Organization/World Health Organization Joint Expert Committee in Guidelines for Evaluation of Probiotics in Food for probiotic lactic acid bacteria drug resistance in 2001. The Oxford Cup method and double-tube dilution method were used to determine the quinolone resistance levels of the isolated strains. Generally, all of the 19 strains showed resistance towards norfloxacin and ciprofloxacin when the Oxford cup method was used, whereas the incidence was lower (to norfloxacin 89.5% and to ciprofloxacin 68.4%) when minimum inhibitory concentration breakpoints (CLSI M100-S23) were tested. Furthermore, gene sequencing was conducted on gyrA and parC of topoisomerase II of these isolated strains. The genetic basis for quinolone resistance may be closely related to mutations in gyrA genes as there were 10 mutation sites in amino-acid sequences encoded by gyrA genes in 10 quinolone resistance strains and 14 mutation sites in Enterococcus durans HZ28, whereas no typical mutations were detected in parC genes.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Hummel, A. S., Hertel, C., Holzapfel, W. H. & Franz, C. M. Antibiotic resistances of starter and probiotic strains of lactic acid bacteria. Appl. Environ. Microbiol. 73, 730–739 (2007).
Caplice, E. & Fitzgerald, G. F. Food fermentations: role of microorganisms in food production and preservation. Int. J. Food Microbiol. 50, 131–149 (1999).
Leroy, F. & De Vuyst, L. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci. 15, 67–78 (2004).
Wood, B. J. & Holzapfel, W. H. N. (eds) in The Genera of Lactic Acid Bacteria, Vol. 2, Springer (Blackie Academic and Professional): London, UK, (1995).
Carr, F. J., Chill, D. & Maida, N. The lactic acid bacteria: a literature survey. Crit. Rev. Microbiol. 28, 281–370 (2002).
Chorostowska-Wynimko, J., Krotkiewski, M., Radomska-Leśniewska, D., Sokolnicka, I. & Skopińska-Rózewska, E. The synergistic effect of lactic acid bacteria and alkylglycerols on humoral immunity in mice. Int. J. Tissue React. 23, 81–87 (2000).
Dàvila, E., Saguer, E., Toldrà, M., Carretero, C. & Parés, D. Preservation of porcine blood quality by means of lactic acid bacteria. Meat Sci. 73, 386–393 (2006).
Shin, H. S., Chung, M. J., Kim, J. E., Lee, K. O. & Ha, N. J. Lactic acid bacteria affect serum cholesterol levels, harmful fecal enzyme activity, and fecal water content. Lipids Health Dis. 8, 21 (2009).
Joint, F. A. O. WHO Working Group Report on Drafting Guidelines for the Evaluation of Probiotics in Food (London, Ontario, Canada, 30 April 2002).
Krasteva, P. V. Molecular engineering: DNA nanoLEGOlogy. Nat. Methods 9, 640–641 (2012).
Méchali, M. Methods in DNA replication. Methods 57, 139 (2012).
Zhang, Y. & Gong, F. DNA repair. Methods 48, 1–2 (2009).
Collin, F., Karkare, S. & Maxwell, A. Exploiting bacterial DNA gyrase as a drug target: current state and perspectives. Appl. Microbiol. Biotechnol. 92, 479–497 (2011).
Xia, R. R., Guo, X. H., Zhang, Y. Z. & Xu., H. Quinolones and the mechanism of quinolone resistance in bacteria. Chin. J. Antibiot. 35, 255–261 (2010).
Hummel, A., Holzapfel, W. H. & Franz, C. M. Characterisation and transfer of antibiotic resistance genes from enterococci isolated from food. Syst. Appl. Microbiol. 30, 1–7 (2007).
Landman, D. & Quale, J. M. Management of infections due to resistant enterococci: a review of therapeutic options. J. Antimicrob. Chemother. 40, 161–170 (1997).
Leclercq, R. Enterococci acquire new kinds of resistance. Clin. Infect. Dis. 24 (Suppl. 1), S80–S84 (1997).
Morrison, D., Woodford, N. & Cookson, B. Enterococci as emerging pathogens of humans. J. Appl. Microbiol. 83, 89S–99S (1997).
Murray, B. E. The life and times of the Enterococcus. Clin. Microbiol. Rev. 3, 46–65 (1990).
Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Third Informational Supplement. M100-S23 (Clinical and Laboratory Standards Institute, 2013).
Kawamura, Y. et al. First Streptococcus agalactiae isolates highly resistant to quinolones, with point mutations in gyrA and parC. Antimicrob. Agents Chemother. 47, 3605–3609 (2003).
Klein, G., Pack, A. & Reuter, G. Antibiotic resistance patterns of enterococci and occurrence of vancomycin-resistant enterococci in raw minced beef and pork in Germany. Appl. Environ. Microbiol. 64, 1825–1830 (1998).
Brisse, S. et al. Association of alterations in ParC and GyrA proteins with resistance of clinical isolates of Enterococcus faecium to nine different fluoroquinolones. Antimicrob. Agents Chemother. 43, 2513–2516 (1999).
Hooper, D. C. Mechanisms of action and resistance of older and newer fluoroquinolones. Clin. Infect. Dis. 31 (Suppl. 2), S24–S28 (2000).
Schmitz., F. J. et al. Characterisation of grlA, grlB, gyrA, and gyrB mutations in 116 unrelated isolates of Staphylococcus aureus and effects of mutations on cipro-floxacin MIC. Antimicrob. Agents Chemother. 42, 1249–1252 (1998).
Pulido, R. P. et al. Resistance to antimicrobial agents in lactobacilli isolated from caper fermentations. Antonie Van Leeuwenhoek. 88, 277–281 (2005).
Zhou, J. S., Pillidge, C. J., Gopal, P. K. & Gill, H. S. Antibiotic susceptibility profiles of new probiotic Lactobacillus and Bifidobacterium strains. Int. J. Food. Microbiol. 98, 211–217 (2005).
European Commission.. Opinion of the Scientific Committee on Animal Nutrition on the criteria for assessing the safety of micro-organisms resistant to antibiotics of human clinical and veterinary importance, European Commission, Health and Consumer Protection Directorate General, Directorate C, Scientific Opinions: Brussels, Belgium, (2002).
FEEDAP, P. Opinion of the scientific panel on additives and products or substances used in animal feed on the updating of the criteria used in assessment of bacteria for resistance to antibiotics of human and veterinary importance. EFSA J. 223, 1–12 (2005).
Zarazaga, M. et al. In vitro activities of ketolide HMR3647, macrolides, and other antibiotics against Lactobacillus, Leuconostoc, and Pediococcus isolates. Antimicrob. Agents Chemother. 43, 3039–3041 (1999).
Fournier, B. & Hooper, D. C. Mutations in topoisomerase IV and DNA gyrase of Staphylococcus aureus: novel pleiotropic effects on quinolone and coumarin activity. Antimicrob. Agents Chemother. 42, 121–128 (1998).
Bébéar, C. M., Charron, A., Bové, J. M., Bébéar, C. & Renaudin, J. Cloning and nucleotide sequences of the Topoisomerase IV parC and parE genes of Mycoplasma hominis. Antimicrob. Agents Chemother. 42, 2024–2031 (1998).
Patel, S. N. et al. Characterization of the quinolone resistant determining regions in clinical isolates of pneumococci collected in Canada. Ann. Clin. Microbiol. Antimicrob. 9, 3 (2010).
Cabral, J. H. M. et al. Crystal structure of the breakage–reunion domain of DNA gyrase. Nature 388, 903–906 (1997).
Gillespie, S. H., Voelker, L. L., Ambler, J. E., Traini, C. & Dickens, A. Fluoroquinolone resistance in Streptococcus pneumoniae: evidence that gyrA mutations arise at a lower rate and that mutation in gyrA or parC predisposes to further mutation. Microb. Drug Resist. 9, 17–24 (2003).
Li, X. Z. & Nikaido, H. Efflux-mediated drug resistance in bacteria. Drugs 64, 159–204 (2004).
Li, X. Z. & Nikaido, H. Efflux-mediated drug resistance in bacteria. Drugs 69, 1555–1623 (2009).
Acknowledgements
We thank the National Natural Science Foundation of China (No. 31060014) for supporting this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Li, S., Li, Z., Wei, W. et al. Association of mutation patterns in GyrA and ParC genes with quinolone resistance levels in lactic acid bacteria. J Antibiot 68, 81–87 (2015). https://doi.org/10.1038/ja.2014.113
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ja.2014.113
This article is cited by
-
Antibiotic resistance in potential probiotic lactic acid bacteria of fermented foods and human origin from Nigeria
BMC Microbiology (2023)
-
Antibiotic Resistance Characterization of Bacteria Isolated from Traditional Chinese Paocai
Current Microbiology (2021)
-
Exploring Probiotic Activity of Lactobacillus sp. Isolated from Indigenous Breeds of Cattle Milk and Fecal Samples in Bhatan Village, MH., IN
Current Microbiology (2020)
-
Antibiotic Resistance of LACTOBACILLUS Strains
Current Microbiology (2019)