Abstract
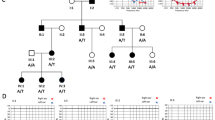
Hereditary hearing impairment (HI) displays extensive genetic heterogeneity. To date, 67 autosomal recessive nonsyndromic hearing impairment (ARNSHI) loci have been mapped, and 24 genes have been identified. This report describes three large consanguineous ARNSHI Pakistani families, all of which display linkage to marker loci located in the genetic interval of DFNB49 locus on chromosome 5q13. Recently, Riazuddin et al. (Am J Hum Genet 2006; 79:1040–1051) reported that variants within the TRIC gene, which encodes tricellulin, are responsible for HI due to DFNB49. TRIC gene sequencing in these three families led to the identification of a novel mutation (IVS4 + 1G > A) in one family and the discovery of a previously described mutation (IVS4 + 2T > C) in two families. It is estimated that 1.06% (95% confidence interval 0.02–3.06%) of families with ARNSHI in Pakistan manifest HI due to mutations in the TRIC gene.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Ando-Akatsuka Y, Saitou M, Hirase T, Kishi M, Sakakibara A, Itoh M, Yonemura S, Furuse M, Tsukita S (1996) Interspecies diversity of the occludin sequence: cDNA cloning of human, mouse, dog, and rat-kangaroo homologues. J Cell Biol 133:43–47
Cottingham R, Indury RM, Schaffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263
Furuse M, Hirase T, Itoh M, Nagafuchi A, Yonemura S, Tsukita S, Tsukita S (1993) Occludin: a novel integral membrane protein localizing at tight junctions. J Cell Biol 123:1777–1788
Gudbjartsson DF, Thorvaldsson T, Kong A, Gunnarsson G, Ingolfsdottir A (2005) Allegro version 2. Nat Genet 37:1015–1016
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp 41:95–98
Ikenouchi J, Furuse M, Furuse K, Sasaki H, Tsukita S, Tsukita S (2005) Tricellulin constitutes a novel barrier at tricellular contacts of epithelial cells. J Cell Biol 171:939–945
Kitajiri S, Miyamoto T, Mineharu A, Sonoda N, Furuse K, Hata M, Sasaki H, Mori Y, Kubota T, Ito J, Furuse M, Tsukita S (2004) Compartmentalization established by claudin-11-based tight junctions in stria vascularis is required for hearing through generation of endocochlear potential. J Cell Sci 117:5087–5096
Kong X, Murphy K, Raj T, He C, White PS, Matise TC (2004) A combined linkage-physical map of the human genome. Am J Hum Genet 75:1143–1148
Mehl AL, Thomson V (1998) Newborn hearing screening: the great omission. Pediatrics 101:e4
Mehl AL, Thomson V (2002) The Colorado newborn hearing screening project, 1992–1999: on the threshold of effective population-based universal newborn hearing screening. Pediatrics 109:e7
Morton NE (1991) Genetic epidemiology of hearing impairment. Ann NY Acad Sci 630:16–31
O’Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266
Petit C (1996) Genes responsible for human hereditary deafness: symphony of a thousand. Nat Genet 14:385–391
Ramzan K, Shaikh RS, Ahmad J, Khan SN, Riazuddin S, Ahmed ZM, Friedman TB, Wilcox ER, Riazuddin S (2005) A new locus for nonsyndromic deafness DFNB49 maps to chromosome 5q12.3-q14.1. Hum Genet 116:17–22
Riazuddin S, Ahmed ZM, Fanning AS, Lagziel A, Kitajiri S, Ramzan K, Khan SN, Chattaraj P, Friedman PL, Anderson JM, Belyantseva IA, Forge A, Riazuddin S, Friedman TB (2006) Tricellulin is a tight-junction protein necessary for hearing. Am J Hum Genet 79:1040–1051
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386
Santos RL, Wajid M, Pham TL, Hussan J, Ali G, Ahmad W, Leal SM (2005) Low prevalence of Connexin 26 (GJB2) variants in Pakistani families with autosomal recessive non-syndromic hearing impairment. Clin Genet 67:61–68
Acknowledgments
We thank the patients and other family members for participating in this study. The work presented here was funded by the Higher Education Commission (HEC), Islamabad, Pakistan, and National Institutes of Health (NIH), National Institute of Deafness and other Communication Disorders (NIDCD), Bethesda, MD, USA, grant DC03594. Muhammad Salman Chishti was supported by indigenous Ph.D. fellowships from HEC and NIDCD grant R01-DC03594. Genotyping services were provided by Center for Inherited Disease Research (CIDR) (Bethesda, MD, USA) and the National Heart, Lung and Blood Institute (NHLBI) Mammalian Genotyping Service (Marshfield, WI, USA). CIDR is fully funded through a federal contract from the NIH to The Johns Hopkins University (Baltimore, MD, USA), contract number N01-HG-65403.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chishti, M.S., Bhatti, A., Tamim, S. et al. Splice-site mutations in the TRIC gene underlie autosomal recessive nonsyndromic hearing impairment in Pakistani families. J Hum Genet 53, 101–105 (2008). https://doi.org/10.1007/s10038-007-0209-3
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1007/s10038-007-0209-3
Keywords
This article is cited by
-
FOXO3/TGF-β signal-dependent ciliogenesis and cell functions during differentiation of temperature-sensitive mouse cochlear precursor hair cells
Histochemistry and Cell Biology (2022)
-
The intestinal epithelial barrier: a therapeutic target?
Nature Reviews Gastroenterology & Hepatology (2017)
-
Genetic Testing of Non-familial Deaf Patients for CIB2 and GJB2 Mutations: Phenotype and Genetic Counselling
Biochemical Genetics (2017)
-
Deletion of Tricellulin Causes Progressive Hearing Loss Associated with Degeneration of Cochlear Hair Cells
Scientific Reports (2015)
-
Molecular genetics of MARVELD2 and clinical phenotype in Pakistani and Slovak families segregating DFNB49 hearing loss
Human Genetics (2015)