Abstract
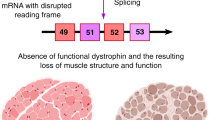
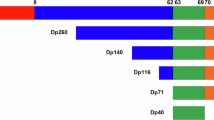
Recent developments in molecular therapies for Duchenne muscular dystrophy (DMD) demand accurate genetic diagnosis, because therapies are mutation specific. The KUCG (Kobe University Clinical Genetics) database for DMD and Becker muscular dystrophy is a hospital-based database comprising 442 cases. Using a combination of complementary DNA (cDNA) and chromosome analysis in addition to conventional genomic DNA-based method, mutation detection was successfully accomplished in all cases, and the largest mutation database of Japanese dystrophinopathy was established. Among 442 cases, deletions and duplications encompassing one or more exons were identified in 270 (61%) and 38 (9%) cases, respectively. Nucleotide changes leading to nonsense mutations or disrupting a splice site were identified in 69 (16%) or 24 (5%) cases, respectively. Small deletion/insertion mutations were identified in 34 (8%) cases. Remarkably, two retrotransposon insertion events were also identified. Dystrophin cDNA analysis successfully revealed novel transcripts with a pseudoexon created by a single-nucleotide change deep within an intron in four cases. X-chromosome abnormalities were identified in two cases. The reading frame rule was upheld for 93% of deletion and 66% of duplication mutation cases. For the application of molecular therapies, induction of exon skipping was deemed the first priority for dystrophinopathy treatment. At one Japanese referral center, the hospital-based mutation database of the dystrophin gene was for the first time established with the highest levels of quality and patient's number.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
References
Zatz, M., Rapaport, D., Vainzof, M., Passos-Bueno, M. R., Bortolini, E. R., Pavanello, R. C. M. et al. Serum creatine-kinase (CK) and pyruvate-kinase activities in Duchenne (DMD) as compared with Becker (BMD) muscular dystrophy. J. Neurol. Sci. 102, 190–196 (1991).
Ahn, A. H. & Kunkel, L. M. The structural and functional diversity of dystrophin. Nat. Genet. 3, 283–291 (1993).
Chamberlain, J. S., Gibbs, R. A., Ranier, J. E., Nguyen, P. N. & Caskey, C. T. Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res. 16, 11141–11156 (1988).
Beggs, A. H., Koenig, M., Boyce, F. M. & Kunkel, L. M. Detection of 98% of DMD/BMD gene deletions by polymerase chain reaction. Hum. Genet. 86, 45–48 (1990).
Monaco, A. P., Bertelson, C. J., Liechti-Gallati, S., Moser, H. & Kunkel, L. M. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics 2, 90–95 (1988).
White, S., Kalf, M., Liu, Q., Villerius, M., Engelsma, D., Kriek, M. et al. Comprehensive detection of genomic duplications and deletions in the DMD gene, by use of multiplex amplifiable probe hybridization. Am. J. Hum. Genet. 71, 365–374 (2002).
Gatta, V., Scarciolla, O., Gaspari, A. R., Palka, C., De Angelis, M. V., Di Muzio, A. et al. Identification of deletions and duplications of the DMD gene in affected males and carrier females by multiple ligation probe amplification (MLPA). Hum. Genet. 117, 92–98 (2005).
Lalic, T., Vossen, R., Coffa, J., Schouten, J., Guc-Scekic, M., Radivojevic, D. et al. Deletion and duplication screening in the DMD gene using MLPA. Eur. J. Hum. Genet. 13, 1231–1234 (2005).
Janssen, B., Hartmann, C., Scholz, V., Jauch, A. & Zschocke, J. MLPA analysis for the detection of deletions, duplications and complex rearrangements in the dystrophin gene: potential and pitfalls. Neurogenetics 6, 29–35 (2005).
Stockley, T. L., Akber, S., Bulgin, N. & Ray, P. N. Strategy for comprehensive molecular testing for Duchenne and Becker muscular dystrophies. Genet. Test. 10, 229–243 (2006).
Zeng, F., Ren, Z., Huang, S., Kalf, M., Mommersteeg, M., Smit, M. et al. Array-MLPA: comprehensive detection of deletions and duplications and its application to DMD patients. Hum. Mutat. 29, 190–197 (2008).
Hegde, M. R., Chin, E. L., Mulle, J. G., Okou, D. T., Warren, S. T. & Zwick, M. E. Microarray-based mutation detection in the dystrophin gene. Hum. Mutat. 29, 1091–1099 (2008).
Takeshima, Y., Yagi, M., Wada, H., Ishibashi, K., Nishiyama, A., Kakumoto, M. et al. Intravenous infusion of an antisense oligonucleotide results in exon skipping in muscle dystrophin mRNA of Duchenne muscular dystrophy. Pediatr. Res. 59, 690–694 (2006).
van Deutekom, J., Janson, A., Ginjaar, I., Frankhuizen, W., Aartsma-Rus, A., Bremmer-Bout, M. et al. Local dystrophin restoration with antisense oligonucleotide PRO051. N. Engl. J. Med. 357, 2677–2686 (2007).
Welch, E. M., Barton, E. R., Zhuo, J., Tomizawa, Y., Friesen, W. J., Trifills, P. et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature 447, 87–91 (2007).
Matsuo, M., Masumura, T., Nishio, H., Nakajima, T., Kitoh, Y., Takumi, T. et al. Exon skipping during splicing of dystrophin mRNA precursor due to an intraexon deletion in the dystrophin gene of Duchenne muscular dystrophy Kobe. J. Clin. Invest. 87, 2127–2131 (1991).
Shiga, N., Takeshima, Y., Sakamoto, H., Inoue, K., Yokota, Y., Yokoyama, M. et al. Disruption of the splicing enhancer sequence within exon 27 of the dystrophin gene by a nonsense mutation induces partial skipping of the exon and is responsible for Becker muscular dystrophy. J. Clin. Invest. 100, 2204–2210 (1997).
Adachi, K., Takeshima, Y., Wada, H., Yagi, M., Nakamura, H. & Matsuo, M. Heterogous dystrophin mRNAs produced by a novel splice acceptor site mutation in intermediate dystrophinopathy. Pediatr. Res. 53, 125–131 (2003).
Matsuo, M., Masumura, T., Nakajima, T., Kitoh, Y., Takumi, T., Nishio, H. et al. A very small frame-shifting deletion within exon 19 of the Duchenne muscular dystrophy gene. Biochem. Biophys. Res. Commun. 170, 963–967 (1990).
Okizuka, Y., Takeshima, Y., Awano, H., Zhang, Z., Yagi, M. & Matsuo, M. Small mutations detected by multiplex ligation-dependant probe amplification of the dystrophin gene. Genet. Test Mol. Biomarkers 13, 427–431 (2009).
Patria, S. Y., Takeshima, Y., Suminaga, R., Nakamura, H., Iwasaki, R., Minagawa, T. et al. A simple explanation for a case of incompatibility with the reading frame theory in Duchenne muscular dystrophy: failure to detect an aberrant restriction fragment in Southern blot analysis. Brain Dev. 21, 386–389 (1999).
Koenig, M., Hoffman, E. P., Bertelson, C. J., Monaco, A. P., Feener, C. & Kunkel, L. M. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell 50, 509–517 (1987).
Tran, V. K., Takeshima, Y., Zhang, Z., Yagi, M., Nishiyama, A., Habara, Y. et al. Splicing analysis disclosed a determinant single nucleotide for exon skipping caused by a novel intra-exonic four-nucleotide deletion in the dystrophin gene. J. Med. Genet. 43, 924–930 (2006).
Tran, V. K., Takeshima, Y., Zhang, Z., Habara, Y., Haginoya, K., Nishiyama, A. et al. A nonsense mutation-created intraexonic splice site is active in the lymphocytes, but not in the skeletal muscle of a DMD patient. Hum. Genet. 120, 737–742 (2007).
Roberts, R. G., Barby, T. F., Manners, E., Bobrow, M. & Bentley, D. R. Direct detection of dystrophin gene rearrangements by analysis of dystrophin mRNA in peripheral blood lymphocytes. Am. J. Hum. Genet. 49, 298–310 (1991).
Patria, S. Y., Alimsardjono, H., Nishio, H., Takeshima, Y., Nakamura, H. & Matsuo, M. A case of Becker muscular dystrophy resulting from the skipping of four contiguous exons (71-74) of the dystrophin gene during mRNA maturation. Proc. Assoc. Am. Phys. 108, 308–314 (1996).
Zhang, Z., Takeshima, Y., Awano, H., Nishiyama, A., Okizuka, Y., Yagi, M. et al. Tandem duplications of two separate fragments of the dystrophin gene in a patient with Duchenne muscular dystrophy. J. Hum. Genet. 53, 215–219 (2008).
Buzin, C. H., Feng, J., Yan, J., Scaringe, W., Liu, Q., den Dunnen, J. et al. Mutation rates in the dystrophin gene: a hotspot of mutation at a CpG dinucleotide. Hum. Mutat. 25, 177–188 (2005).
Tuffery-Giraud, S., Beroud, C., Leturcq, F., Yaou, R. B., Hamroun, D., Michel-Calemard, L. et al. Genotype-phenotype analysis in 2405 patients with a dystrophinopathy using the UMD-DMD database: a model of nationwide knowledgebase. Hum. Mut. 30, 934–945 (2009).
Nishiyama, A., Takeshima, Y., Zhang, Z., Habara, Y., Tran, T. H., Yagi, M. et al. Dystrophin nonsense mutations can generate alternative rescue transcripts in lymphocytes. Ann. Hum. Genet. 72, 717–724 (2008).
Suminaga, R., Takeshima, Y., Wada, H., Yagi, M. & Matsuo, M. C-terminal truncated dystrophin identified in skeletal muscle of an asymptomatic boy with a novel nonsense mutation of the dystrophin gene. Pediatr. Res. 56, 739–743 (2004).
Yagi, M., Takeshima, Y., Wada, H., Nakamura, H. & Matsuo, M. Two alternative exons can result from activation of the cryptic splice acceptor site deep within intron 2 of the dystrophin gene in a patient with as yet asymptomatic dystrophinopathy. Hum. Genet. 112, 164–170 (2003).
Habara, Y., Takeshima, Y., Awano, H., Okizuka, Y., Zhang, Z., Saiki, K. et al. In vitro splicing analysis reveals that availability of a cryptic splice site is not a determinant for alternative splicing patterns caused by +1G>A mutations in introns of the dystrophin gene. J. Med. Genet. 46, 542–547 (2009).
Narita, N., Nishio, H., Kitoh, Y., Ishikawa, Y., Ishikawa, Y., Minami, R. et al. Insertion of a 5′ truncated L1 element into the 3′ end of exon 44 of the dystrophin gene resulted in skipping of the exon during splicing in a case of Duchenne muscular dystrophy. J. Clin. Invest. 91, 1862–1867 (1993).
Trimarco, A., Torella, A., Piluso, G., Maria Ventriglia, V., Politano, L. & Nigro, V. Log-PCR: a new tool for immediate and cost-effective diagnosis of up to 85% of dystrophin gene mutations. Clin. Chem. 54, 973–981 (2008).
Taylor, P. J., Maroulis, S., Mullan, G. L., Pedersen, R. L., Baumli, A., Elakis, G. et al. Measurement of the clinical utility of a combined mutation detection protocol in carriers of Duchenne and Becker muscular dystrophy. J. Med. Genet. 44, 368–372 (2007).
Ashton, E. J., Yau, S. C., Deans, Z. C. & Abbs, S. J. Simultaneous mutation scanning for gross deletions, duplications and point mutations in the DMD gene. Eur. J. Hum. Genet. 16, 53–61 (2008).
Aartsma-Rus, A., Van Deutekom, J. C., Fokkema, I. F., Van Ommen, G. J. & Den Dunnen, J. T. Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve 34, 135–144 (2006).
Hwa, H. L., Chang, Y. Y., Chen, C. H., Kao, Y. S., Jong, Y. J., Chao, M. C. et al. Multiplex ligation-dependent probe amplification identification of deletions and duplications of the Duchenne muscular dystrophy gene in Taiwanese subjects. J. Formos. Med. Assoc. 106, 339–346 (2007).
Schwartz, M. & Duno, M. Improved molecular diagnosis of dystrophin gene mutations using the multiplex ligation-dependent probe amplification method. Genet. Test. 8, 361–367 (2004).
Gurvich, O. L., Tuohy, T. M., Howard, M. T., Finkel, R. S., Medne, L., Anderson, C. B. et al. DMD pseudoexon mutations: splicing efficiency, phenotype, and potential therapy. Ann. Neurol. 63, 81–89 (2008).
Hwa, H. L., Chang, Y. Y., Huang, C. H., Chen, C. H., Kao, Y. S., Jong, Y. J. et al. Small mutations of the DMD gene in Taiwanese families. J. Formos. Med. Assoc. 107, 463–469 (2008).
Musova, Z., Hedvicakova, P., Mohrmann, M., Tesarova, M., Krepelova, A., Zeman, J. et al. A novel insertion of a rearranged L1 element in exon 44 of the dystrophin gene: further evidence for possible bias in retroposon integration. Biochem. Biophys. Res. Commun. 347, 145–149 (2006).
Matsuo, M. Duchenne/Becker muscular dystrophy: from molecular diagnosis to gene therapy. Brain Dev. 18, 167–172 (1996).
Acknowledgements
This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan, a Health and Labor Science Research Grant (Research on Psychiatric and Neurological Diseases and Mental Health) and a Research Grant for Nervous and Mental Disorders from the Ministry of Health, Labor and Welfare.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Takeshima, Y., Yagi, M., Okizuka, Y. et al. Mutation spectrum of the dystrophin gene in 442 Duchenne/Becker muscular dystrophy cases from one Japanese referral center. J Hum Genet 55, 379–388 (2010). https://doi.org/10.1038/jhg.2010.49
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/jhg.2010.49
Keywords
This article is cited by
-
NGS-based targeted sequencing identified six novel variants in patients with Duchenne/Becker muscular dystrophy from southwestern China
BMC Medical Genomics (2023)
-
Benchmarking splice variant prediction algorithms using massively parallel splicing assays
Genome Biology (2023)
-
Cell-Based and Gene-Based Therapy Approaches in Neuro-orthopedic Disorders: a Literature Review
Regenerative Engineering and Translational Medicine (2023)
-
Small mutations in Duchenne/Becker muscular dystrophy in 164 unrelated Polish patients
Journal of Applied Genetics (2021)
-
Identification of two rare mutations c.1318G>A and c.6438+2T>G in a Chinese DMD family as genetic markers
Genes & Genomics (2020)