Abstract
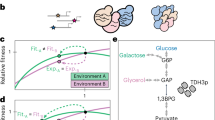
The effects of mutations on phenotype and fitness may depend on the environment (phenotypic plasticity), other mutations (genetic epistasis) or both. Here we examine the fitness effects of 18 random insertion mutations in E. coli in two resource environments and five genetic backgrounds. We tested each mutation for plasticity and epistasis by comparing its fitness effects across these ecological and genetic contexts. Some mutations had no measurable effect in any of these contexts. None of the mutations had effects on phenotypic plasticity that were independent of genetic background. However, half the mutations had epistatic interactions such that their effects differed among genetic backgrounds, usually in an environment-dependent manner. Also, the pattern of mutational effects across backgrounds indicated that epistasis had been shaped primarily by unique events in the evolutionary history of a population rather than by repeatable events associated with shared environmental history.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Wade, M.J. Epistasis as a genetic constraint within populations and an accelerant of adaptive divergence among them. in Epistasis and the Evolutionary Process (eds. Wolf, J.B., Brodie, E.D.I. & Wade, M.J.) 213–231 (Oxford Univ. Press, Oxford, UK, 2000).
Wade, M.J. & Goodnight, C.J. The theories of Fisher and Wright in the context of metapopulations: when nature does many small experiments. Evolution 52, 1537–1553 (1998).
Otto, S.P. & Feldman, M.W. Deleterious mutations, variable epistatic interactions, and the evolution of recombination. Theor. Pop. Biol. 51, 134–147 (1997).
Hansen, T.F. & Wagner, G.P. Epistasis and the mutation load: a measurement theoretical approach. Genetics 158, 477–485 (2001).
Kondrashov, F.A. & Kondrashov, A.S. Multidimensional epistasis and the disadvantage of sex. Proc. Natl. Acad. Sci. USA 98, 12089–12092 (2001).
Whitlock, M.C., Phillips, P.C., Moore, F.B.-G. & Tonsor, S.J. Multiple fitness peaks and epistasis. Annu. Rev. Ecol. Syst. 26, 601–629 (1995).
Dean, A.M. A molecular investigation of genotype by environment interactions. Genetics 139, 19–33 (1995).
Fedorowicz, G.M., Fry, J.D., Anholt, R.R. & Mackay, T.F.C. Epistatic interactions between smell-impaired loci in Drosophila melanogaster. Genetics 148, 1885–1891 (1998).
Dykhuizen, D.E. & Hartl, D.L. Functional effects of PGI allozymes in Escherichia coli. Genetics 105, 1–18 (1983).
Lenski, R.E. et al. Epistatic effects of promoter and repressor functions of the Tn10 tetracycline-resistance operon on the fitness of Escherichia coli. Mol. Ecol. 3, 127–135 (1994).
De Visser, J.A.G.M., Hoekstra, R.F. & van den Ende, H. Test of interaction between genetic markers that affect fitness in Aspergillus niger. Evolution 51, 1499–1505 (1996).
Elena, S.F. & Lenski, R.E. Test of synergistic interactions among deleterious mutations in bacteria. Nature 390, 395–398 (1997).
Elena, S.F., Ekunwe, L., Hajela, N., Oden, S.A. & Lenski, R.E. Distribution of fitness effects caused by random insertion mutations in Escherichia coli. Genetica 102/103, 349–358 (1998).
Elena, S.F. & Lenski, R.E. Epistasis between new mutations and genetic background, and a test of genetic canalization. Evolution 55, 1746–1752 (2001).
Remold, S.K. & Lenski, R.E. Contribution of individual random mutations to genotype-by-environment interactions in Escherichia coli. Proc. Natl. Acad. Sci. USA 98, 11388–11393 (2001).
Kishony, R. & Leibler, S. Environmental stresses can alleviate the average deleterious effect of mutations. J. Biol. 2, 14 (2003).
Miller, J.H. A Short Course in Bacterial Genetics (Cold Spring Harbor Laboratory Press, Plainview, New York, USA, 1992).
Kleckner, N., Bender, J. & Gottesman, S. Uses of transposons with emphasis on Tn10. Meth. Enzymol. 204, 139–180 (1991).
Lenski, R.E., Rose, M.R., Simpson, S.C. & Tadler, S.C. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am. Nat. 138, 1315–1341 (1991).
Travisano, M. & Lenski, R.E. Long-term experimental evolution in Escherichia coli. IV. Targets of selection and the specificity of adaptation. Genetics 143, 15–26 (1996).
Travisano, M. Long-term experimental evolution in Escherichia coli. VI. Environmental constraints on adaptation and divergence. Genetics 146, 471–479 (1997).
Schmalhausen, I.I. Factors of Evolution: The Theory of Stabilizing Selection (Blakiston, Philadelphia, USA, 1949).
Podlich, D.W. & Cooper, M. Modelling plant breeding programs as search strategies on a complex response surface. in Simulated Evolution and Learning (eds. McKay, B., Yao, X., Newton, C.S., Kim, J.-H. & Furuhashi, T.) 171–178 (Springer, Canberra, Australia, 1998).
Lenski, R.E. & Travisano, M. Dynamics of adaptation and diversification: a 10,000-generation experiment with bacterial populations. Proc. Natl. Acad. Sci. USA 91, 6808–6814 (1994).
Lenski, R.E., Winkworth, C.L. & Riley, M.A. Rates of DNA sequence evolution in experimental populations of Escherichia coli during 20,000 generations. J. Mol. Evol. 56, 498–508 (2003).
Lenski, R.E. Phenotypic and genomic evolution during a 20,000-generation experiment with the bacterium, Escherichia coli. Plant Breed. Rev. 24, 225–265 (2004).
Levin, B.R., Stewart, F.M. & Chao, L. Resource-limited growth, competition and predation: a model and experimental studies with bacteria and bacteriophage. Am. Nat. 111, 3–24 (1977).
Littell, R.C., Milliken, G.A., Stroup, W.W. & Wolfinger, R.D. SAS System for Mixed Models (SAS Institute, Cary, North Carolina, USA, 1996).
Sokal, R.R. & Rohlf, F.J. Biometry (Freeman, New York, USA, 1995).
Acknowledgements
We thank S. Elena and M. Travisano for sharing strains; N. Hajela for technical support; C. Borland, T. Cooper, H. Eisthen, B. Lundrigan, P. Moore and M. Travisano for valuable comments; and R. Wolfinger and C. Duarte for statistical advice. This work was supported by a fellowship from the US National Institutes of Health to S.K.R. and a grant from the National Science Foundation to R.E.L.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Remold, S., Lenski, R. Pervasive joint influence of epistasis and plasticity on mutational effects in Escherichia coli. Nat Genet 36, 423–426 (2004). https://doi.org/10.1038/ng1324
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ng1324
This article is cited by
-
Allopatric divergence of cooperators confers cheating resistance and limits effects of a defector mutation
BMC Ecology and Evolution (2022)
-
Endocytic recycling protein EHD1 regulates primary cilia morphogenesis and SHH signaling during neural tube development
Scientific Reports (2016)
-
RNAi phenotypes are influenced by the genetic background of the injected strain
BMC Genomics (2013)
-
Evolutionary rescue from extinction is contingent on a lower rate of environmental change
Nature (2013)
-
Magnitude and sign epistasis among deleterious mutations in a positive-sense plant RNA virus
Heredity (2012)