Abstract
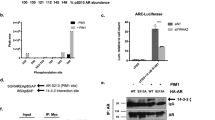
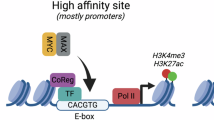
The serine/threonine kinase PIM1, cooperates with MYC in cell cycle progression and tumorigenesis. However, the nature of this cooperation remains elusive. Here we show that PIM1 contributes to the transcriptional activation of MYC-target genes by phosphorylating the histone H3 nucleosome at Serine 10 (H3S10). Recombinant PIM1 directly phosphorylates H3S10 on the nucleosome in vitro. Following growth factor stimulation, PIM1 accumulates in the nucleus where it forms a complex with the MYC/MAX dimer via the MYC BoxII domain (MBII). Immunofluorescence analysis coupled with in vivo run-on shows a high degree of PIM1 and MYC co-localization in the nucleus at sites of active transcription. Expression profile analysis revealed that PIM1 contributes to the regulation of 20% of the MYC-regulated genes. Chromatin immunoprecipitation (ChIP) analysis in MYC silenced cells demonstrates that MYC recruits PIM1 to the E boxes of the MYC-target genes FOSL1 (FRA1) and ID2. PIM1 knockdown as well as the over-expression of the kinase inactive mutant demonstrate that PIM1 is required for H3S10 phosphorylation at FOSL1 and ID2 MYC-binding sites and for their transcriptional activation. Moreover, we show that PIM1-dependent H3S10 phosphorylation contributes to MYC transforming capacity. These results establish a new function for PIM1 as a MYC cofactor that phosphorylates the chromatin at MYC-target loci and suggest that nucleosome phosphorylation, at E boxes, contributes to MYC-dependent transcriptional activation and cellular transformation.
Similar content being viewed by others
Article PDF
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Oliviero, S., Zippo, A., De Robertis, A. et al. PIM1-dependent phosphorylation of Histone H3 at Serine 10 is required for MYC-dependent transcriptional activation and oncogenic transformation.. Nat Prec (2007). https://doi.org/10.1038/npre.2007.100.1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/npre.2007.100.1