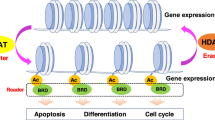
Key Points
-
Epigenetic regulation of gene expression relies on covalent, reversible and sequence-specific modifications of histone proteins that package DNA. Deregulation of this control mechanism can lead to multiple diseases.
-
Acetylation and methylation of lysine and arginine residues are the most abundant modifications of the known histone marks. Histone acetyl marks are written by histone acetyltranferases, read by bromodomain-containing proteins and erased by histone deacetylases.
-
Histone methyl marks are written by protein methyltransferases and read by proteins containing chromodomains, Tudor domains, malignant brain tumour domains, PWWP domains and plant homeodomain (PHD) fingers. Histone methyl marks are erased by lysine demethylases.
-
Domains that write, read or erase histone marks are often grouped within a single gene, and these proteins are often combined within multiprotein complexes that mediate crosstalk between specific marks.
-
When they are mutated, aberrantly expressed or driven by aberrant upstream signals, writers, readers and erasers of histone marks can affect the expression patterns of genes that lead to — or even drive — and maintain disease states. Direct associations have been observed in cancer, neuropsychiatric disorders, inflammation and metabolic diseases.
-
Several histone deacetylase inhibitors have reached the clinic and two are approved in oncology. Small-molecule antagonists of bromodomain-containing proteins have demonstrated efficacy in animal models of cancer and inflammation. The chemical tractability of histone acetyltransferases, however, remains unclear.
-
Potent protein methyltransferase inhibitors have been reported, and one compound was active against tumour xenografts in mice. Lysine demethylase inhibitors and antagonists of methyl-lysine readers are at an earlier development stage.
Abstract
Epigenetic regulation of gene expression is a dynamic and reversible process that establishes normal cellular phenotypes but also contributes to human diseases. At the molecular level, epigenetic regulation involves hierarchical covalent modification of DNA and the proteins that package DNA, such as histones. Here, we review the key protein families that mediate epigenetic signalling through the acetylation and methylation of histones, including histone deacetylases, protein methyltransferases, lysine demethylases, bromodomain-containing proteins and proteins that bind to methylated histones. These protein families are emerging as druggable classes of enzymes and druggable classes of protein–protein interaction domains. In this article, we discuss the known links with disease, basic molecular mechanisms of action and recent progress in the pharmacological modulation of each class of proteins.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Meaney, M. J. Epigenetics and the biological definition of gene × environment interactions. Child Dev. 81, 41–79 (2010).
Kelly, T. K., De Carvalho, D. D. & Jones, P. A. Epigenetic modifications as therapeutic targets. Nature Biotech. 28, 1069–1078 (2010).
Meaney, M. J. & Ferguson-Smith, A. C. Epigenetic regulation of the neural transcriptome: the meaning of the marks. Nature Neurosci. 13, 1313–1318 (2010).
Portela, A. & Esteller, M. Epigenetic modifications and human disease. Nature Biotech. 28, 1057–1068 (2010).
Allis, C. D., Jenuwein, T. & Reinberg, D. Epigenetics (Cold Spring Harbor Laboratory Press, New York, 2007).
Taberlay, P. C. & Jones, P. A. DNA methylation and cancer. Prog. Drug Res. 67, 1–23 (2011).
Strahl, B. D. & Allis, C. D. The language of covalent histone modifications. Nature 403, 41–45 (2000).
Turner, B. M. Histone acetylation and an epigenetic code. Bioessays 22, 836–845 (2000).
Turner, B. M. Cellular memory and the histone code. Cell 111, 285–291 (2002).
Schreiber, S. L. & Bernstein, B. E. Signaling network model of chromatin. Cell 111, 771–778 (2002).
Ernst, J. et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–49 (2011).
Allfrey, V. G., Faulkner, R. & Mirsky, A. E. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl Acad. Sci. USA 51, 786–794 (1964). References 7, 8 and 12 highlight the role of histone modifications in gene regulation, and the concept of an epigenetic code.
Shahbazian, M. D. & Grunstein, M. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76, 75–100 (2007).
Das, C., Lucia, M. S., Hansen, K. C. & Tyler, J. K. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature 459, 113–117 (2009).
Filippakopoulos, P. et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 30 Mar 2012 (doi: 10.1016/j.cell.2012.02.013). This is a full structural analysis of the entire human bromodomain-containing family of proteins, which established the size and details of this target class.
Haynes, S. R. et al. The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res. 20, 2603 (1992).
Tsukada, Y. et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439, 811–816 (2006).
Shi, Y. et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953 (2004). References 17 and 18 describe the discovery of the two classes of lysine demethylases, both of which belong to larger druggable enzyme classes.
Trojer, P. et al. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell 129, 915–928 (2007).
Nielsen, A. L. et al. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol. Cell 7, 729–739 (2001).
Lee, M. G. et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 318, 447–450 (2007).
Kuzmichev, A., Nishioka, K., Erdjument-Bromage, H., Tempst, P. & Reinberg, D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 16, 2893–2905 (2002).
Bannister, A. J. & Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 (2011).
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007).
Di Lorenzo, A. & Bedford, M. T. Histone arginine methylation. FEBS Lett. 585, 2024–2031 (2011).
Kirmizis, A. et al. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature 449, 928–932 (2007).
Guccione, E. et al. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature 449, 933–937 (2007).
Jones, P. A. & Baylin, S. B. The epigenomics of cancer. Cell 128, 683–692 (2007).
Krennhrubec, K., Marshall, B. L., Hedglin, M., Verdin, E. & Ulrich, S. M. Design and evaluation of 'Linkerless' hydroxamic acids as selective HDAC8 inhibitors. Bioorg. Med. Chem. Lett. 17, 2874–2878 (2007).
Oehme, I. et al. Histone deacetylase 8 in neuroblastoma tumorigenesis. Clin. Cancer Res. 15, 91–99 (2009).
Chi, P., Allis, C. D. & Wang, G. G. Covalent histone modifications — miswritten, misinterpreted and mis-erased in human cancers. Nature Rev. Cancer 10, 457–469 (2010). This is a summary of the extensive number and types of 'errors' in epigenetic proteins implicated in cancer.
Simon, J. A. & Lange, C. A. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat. Res. 647, 21–29 (2008).
Okada, Y. et al. hDOT1L links histone methylation to leukemogenesis. Cell 121, 167–178 (2005).
Daigle, S. R. et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell 20, 53–65 (2011). This is the first example of selective inhibition of a histone methyltransferase that has efficacy in a cancer model.
Yap, D. B. et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood 117, 2451–2459 (2010).
Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2011).
Darnell, J. E. Jr. Transcription factors as targets for cancer therapy. Nature Rev. Cancer 2, 740–749 (2002).
Beroukhim, R. et al. The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010).
Meyer, N. & Penn, L. Z. Reflecting on 25 years with MYC. Nature Rev. Cancer 8, 976–990 (2008).
Delmore, J. E. et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917 (2011). References 36 and 40 provide the first examples of bromodomain inhibition that has apparent therapeutic benefit.
Dawson, M. A. et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478, 529–533 (2011).
Zuber, J. et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478, 524–528 (2011).
Kalashnikova, E. V. et al. ANCCA/ATAD2 overexpression identifies breast cancer patients with poor prognosis, acting to drive proliferation and survival of triple-negative cells through control of B-Myb and EZH2. Cancer Res. 70, 9402–9412 (2010).
Caron, C. et al. Functional characterization of ATAD2 as a new cancer/testis factor and a predictor of poor prognosis in breast and lung cancers. Oncogene 29, 5171–5181 (2010).
Zhang, Y. et al. PR-domain-containing Mds1–Evi1 is critical for long-term hematopoietic stem cell function. Blood 118, 3853–3861 (2011).
Velichutina, I. et al. EZH2-mediated epigenetic silencing in germinal center B cells contributes to proliferation and lymphomagenesis. Blood 116, 5247–5255 (2010).
Trubia, M. et al. Characterization of a recurrent translocation t(2;3)(p15–22;q26) occurring in acute myeloid leukaemia. Leukemia 20, 48–54 (2006).
Wang, G. G. et al. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature 459, 847–851 (2009).
Parsons, D. W. et al. The genetic landscape of the childhood cancer medulloblastoma. Science 331, 435–439 (2011).
Northcott, P. A. et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nature Genet. 41, 465–472 (2009).
Wang, X. & Jin, H. The epigenetic basis of the Warburg effect. Epigenetics 5, 566–568 (2010).
Gorrini, C. et al. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature 448, 1063–1067 (2007).
Bouwman, P. et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nature Struct. Mol. Biol. 17, 688–695 (2010).
Williams, S. R. et al. Haploinsufficiency of HDAC4 causes brachydactyly mental retardation syndrome, with brachydactyly type E, developmental delays, and behavioral problems. Am. J. Hum. Genet. 87, 219–228 (2010).
Wang, J. et al. CBP histone acetyltransferase activity regulates embryonic neural differentiation in the normal and Rubinstein–Taybi syndrome brain. Dev. Cell 18, 114–125 (2010).
Chen, G., Zou, X., Watanabe, H., van Deursen, J. M. & Shen, J. CREB binding protein is required for both short-term and long-term memory formation. J. Neurosci. 30, 13066–13077 (2010).
Schaefer, A. et al. Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron 64, 678–691 (2009).
Kleefstra, T. et al. Further clinical and molecular delineation of the 9q subtelomeric deletion syndrome supports a major contribution of EHMT1 haploinsufficiency to the core phenotype. J. Med. Genet. 46, 598–606 (2009).
Kramer, J. M. & van Bokhoven, H. Genetic and epigenetic defects in mental retardation. Int. J. Biochem. Cell Biol. 41, 96–107 (2009).
Chiurazzi P., Schwartz C. E., Gecz, J. & Neri G. XLMR genes: update 2007. Eur. J. Hum. Genet. 16, 422–434 (2008).
Kleine-Kohlbrecher, D. et al. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol. Cell 38, 165–178 (2010).
Zakhary, S. M. et al. Distribution analysis of deacetylase SIRT1 in rodent and human nervous systems. Anat. Rec. (Hoboken) 293, 1024–1032 (2010).
Janssen, C. et al. Differential histone deacetylase mRNA expression patterns in amyotrophic lateral sclerosis. J. Neuropathol. Exp. Neurol. 69, 573–581 (2010).
Riessland, M. et al. SAHA ameliorates the SMA phenotype in two mouse models for spinal muscular atrophy. Hum. Mol. Genet. 19, 1492–1506 (2010).
Covington, H. E. et al. Antidepressant actions of histone deacetylase inhibitors. J. Neurosci. 29, 11451–11460 (2009).
Loe-Mie, Y. et al. SMARCA2 and other genome-wide supported schizophrenia-associated genes: regulation by REST/NRSF, network organization and primate-specific evolution. Hum. Mol. Genet. 19, 2841–2857 (2010).
Severinsen, J. E. et al. Evidence implicating BRD1 with brain development and susceptibility to both schizophrenia and bipolar affective disorder. Mol. Psychiatry 11, 1126–1138 (2006).
Lin, H. S. et al. Anti-rheumatic activities of histone deacetylase (HDAC) inhibitors in vivo in collagen-induced arthritis in rodents. Br. J. Pharmacol. 150, 862–872 (2007).
Grabiec, A. M., Korchynskyi, O., Tak, P. P. & Reedquist, K. A. Histone deacetylase inhibitors suppress rheumatoid arthritis fibroblast-like synoviocyte and macrophage IL-6 production by accelerating mRNA decay. Ann. Rheum. Dis. 71, 424–431 (2011).
Shakespear, M. R., Halili, M. A., Irvine, K. M., Fairlie, D. P. & Sweet, M. J. Histone deacetylases as regulators of inflammation and immunity. Trends Immunol. 32, 335–343 (2011).
Beier, U. H., Akimova, T., Liu, Y., Wang, L. & Hancock, W. W. Histone/protein deacetylases control Foxp3 expression and the heat shock response of T-regulatory cells. Curr. Opin. Immunol. 23, 670–678 (2011).
de Zoeten, E. F. et al. Histone deacetylase 6 and heat shock protein 90 control the functions of Foxp3+ T-regulatory cells. Mol. Cell Biol. 31, 2066–2078 (2011).
Galli, M., Van Gool, F. & Leo, O. Sirtuins and inflammation: friends or foes? Biochem. Pharmacol. 81, 569–576 (2011).
Yoshizaki, T. et al. SIRT1 inhibits inflammatory pathways in macrophages and modulates insulin sensitivity. Am. J. Physiol. Endocrinol. Metab. 298, e419–e428 (2010).
Ghizzoni, M., Haisma, H. J., Maarsingh, H. & Dekker, F. J. Histone acetyltransferases are crucial regulators in NF-κB mediated inflammation. Drug Discov. Today 16, 504–511 (2011).
Nicodeme, E. et al. Suppression of inflammation by a synthetic histone mimic. Nature 468, 1119–1123 (2010).
Chen, X., El Gazzar, M., Yoza, B. K. & McCall, C. E. The NF-κB factor RelB and histone H3 lysine methyltransferase G9a directly interact to generate epigenetic silencing in endotoxin tolerance. J. Biol. Chem. 284, 27857–27865 (2009).
van Essen, D., Zhu, Y. & Saccani, S. A feed-forward circuit controlling inducible NF-κB target gene activation by promoter histone demethylation. Mol. Cell 39, 750–760 (2010).
De Santa, F. et al. Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. EMBO J. 28, 3341–3352 (2009).
Ishii, M. et al. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood 114, 3244–3254 (2009).
Ciavatta, D. J. et al. Epigenetic basis for aberrant upregulation of autoantigen genes in humans with ANCA vasculitis. J. Clin. Invest. 120, 3209–3219 (2010).
Hu, N. et al. Abnormal histone modification patterns in lupus CD4+ T cells. J. Rheumatol. 35, 804–810 (2008).
Zhong, L. & Mostoslavsky, R. Fine tuning our cellular factories: sirtuins in mitochondrial biology. Cell Metab. 13, 621–626 (2011).
Zillikens, M. C. et al. SIRT1 genetic variation is related to BMI and risk of obesity. Diabetes 58, 2828–2834 (2009).
Cardellini, M. et al. TIMP3 is reduced in atherosclerotic plaques from subjects with type 2 diabetes and increased by SirT1. Diabetes 58, 2396–2401 (2009).
Orimo, M. et al. Protective role of SIRT1 in diabetic vascular dysfunction. Arterioscler. Thromb. Vasc. Biol. 29, 889–894 (2009).
Vetterli, L., Brun, T., Giovannoni, L., Bosco, D. & Maechler, P. Resveratrol potentiates glucose-stimulated insulin secretion in INS-1E β-cells and human islets through a SIRT1-dependent mechanism. J. Biol. Chem. 286, 6049–6060 (2011).
Ramadori, G. et al. Central administration of resveratrol improves diet-induced diabetes. Endocrinology 150, 5326–5333 (2009).
Fischer-Posovszky, P. et al. Resveratrol regulates human adipocyte number and function in a Sirt1-dependent manner. Am. J. Clin. Nutr. 92, 5–15 (2010).
Brasacchio, D. et al. Hyperglycemia induces a dynamic cooperativity of histone methylase and demethylase enzymes associated with gene-activating epigenetic marks that coexist on the lysine tail. Diabetes 58, 1229–1236 (2009).
Siebel, A. L., Fernandez, A. Z. & El-Osta, A. Glycemic memory associated epigenetic changes. Biochem. Pharmacol. 80, 1853–1859 (2010).
Meissner, A. Epigenetic modifications in pluripotent and differentiated cells. Nature Biotech. 28, 1079–1088 (2010).
Borowiak, M. et al. Small molecules efficiently direct endodermal differentiation of mouse and human embryonic stem cells. Cell Stem Cell 4, 348–358 (2009).
Hsieh, J., Nakashima, K., Kuwabara, T., Mejia, E. & Gage, F. H. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc. Natl Acad. Sci. USA 101, 16659–16664 (2004).
Hao, Y. et al. Mood stabilizer valproate promotes ERK pathway-dependent cortical neuronal growth and neurogenesis. J. Neurosci. 24, 6590–6599 (2004).
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006).
Huangfu, D. et al. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nature Biotech. 26, 1269–1275 (2008).
Shi, Y. et al. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell 2, 525–528 (2008).
de Ruijter, A. J., van Gennip, A. H., Caron, H. N., Kemp, S. & van Kuilenburg, A. B. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 370, 737–749 (2003).
Finnin, M. S. et al. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature 401, 188–193 (1999).
Sauve, A. A. Sirtuin chemical mechanisms. Biochim. Biophys. Acta 1804, 1591–1603 (2010).
Sauve, A. A., Wolberger, C., Schramm, V. L. & Boeke, J. D. The biochemistry of sirtuins. Annu. Rev. Biochem. 75, 435–465 (2006).
Choudhary, C. et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 (2009).
Kovacs, J. J. et al. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol. Cell 18, 601–607 (2005).
Hubbert, C. et al. HDAC6 is a microtubule-associated deacetylase. Nature 417, 455–458 (2002).
Prince, H. M., Bishton, M. J. & Harrison, S. J. Clinical studies of histone deacetylase inhibitors. Clin. Cancer Res. 15, 3958–3969 (2009).
Grant, S., Easley, C. & Kirkpatrick, P. Vorinostat. Nature Rev. Drug Discov. 6, 21–22 (2007).
Marks, P. A. The clinical development of histone deacetylase inhibitors as targeted anticancer drugs. Expert Opin. Investig. Drugs 19, 1049–1066 (2010).
Butler, L. M. et al. Suberoylanilide hydroxamic acid, an inhibitor of histone deacetylase, suppresses the growth of prostate cancer cells in vitro and in vivo. Cancer Res. 60, 5165–5170 (2000). This was the first evidence that an HDAC inhibitor has anticancer activity in an animal model.
Schapira, M. Structural biology of human metal-dependent histone deacetylases. Handb. Exp. Pharmacol. 206, 225–240 (2011).
Bradner, J. E. et al. Chemical phylogenetics of histone deacetylases. Nature Chem. Biol. 6, 238–243 (2010).
Lahm, A. et al. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc. Natl Acad. Sci. USA 104, 17335–17340 (2007).
Schuetz, A. et al. Human HDAC7 harbors a class IIa histone deacetylase-specific zinc binding motif and cryptic deacetylase activity. J. Biol. Chem. 283, 11355–11363 (2008).
Mihaylova, M. M. et al. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell 145, 607–621 (2011).
Bantscheff, M. et al. Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nature Biotech. 29, 255–265 (2011).
Vaquero, A. et al. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 20, 1256–1261 (2006).
Vaquero, A. et al. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol. Cell 16, 93–105 (2004).
Vempati, R. K. et al. p300-mediated acetylation of histone H3 lysine 56 functions in DNA damage response in mammals. J. Biol. Chem. 285, 28553–28564 (2010).
Michishita, E. et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452, 492–496 (2008).
Du, J. et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334, 806–809 (2011).
Liu, B. et al. Identification and characterization of propionylation at histone H3 lysine 23 in mammalian cells. J. Biol. Chem. 284, 32288–32295 (2009).
Vaziri, H. et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107, 149–159 (2001).
Haigis, M. C. & Sinclair, D. A. Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 5, 253–295 (2010).
Milne, J. C. et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature 450, 712–716 (2007).
Stunkel, W. & Campbell, R. M. Sirtuin 1 (SIRT1): the misunderstood HDAC. J. Biomol. Screen. 16, 1153–1169 (2011).
Pacholec, M. et al. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J. Biol. Chem. 285, 8340–8351 (2010).
Solomon, J. M. et al. Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol. Cell Biol. 26, 28–38 (2006).
Liu, X. et al. The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature 451, 846–850 (2008).
Piaz, F. D. et al. Chemical biology of histone acetyltransferase natural compounds modulators. Mol. Divers. 15, 401–416 (2011).
Dekker, F. J. & Haisma, H. J. Histone acetyl transferases as emerging drug targets. Drug Discov. Today 14, 942–948 (2009).
Wisastra, R. et al. Isothiazolones; thiol-reactive inhibitors of cysteine protease cathepsin B and histone acetyltransferase PCAF. Org. Biomol. Chem. 9, 1817–1822 (2011).
Lau, O. D. et al. HATs off: selective synthetic inhibitors of the histone acetyltransferases p300 and PCAF. Mol. Cell 5, 589–595 (2000).
Bowers, E. M. et al. Virtual ligand screening of the p300/CBP histone acetyltransferase: identification of a selective small molecule inhibitor. Chem. Biol. 17, 471–482 (2010).
Santer, F. R. et al. Inhibition of the acetyltransferases p300 and CBP reveals a targetable function for p300 in the survival and invasion pathways of prostate cancer cell lines. Mol. Cancer Ther. 10, 1644–1655 (2011).
Spannhoff, A., Hauser, A. T., Heinke, R., Sippl, W. & Jung, M. The emerging therapeutic potential of histone methyltransferase and demethylase inhibitors. ChemMedChem 4, 1568–1582 (2009).
Copeland, R. A., Solomon, M. E. & Richon, V. M. Protein methyltransferases as a target class for drug discovery. Nature Rev. Drug Discov. 8, 724–732 (2009).
Kubicek, S. et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol. Cell 25, 473–481 (2007). This was the first example of selective, substrate-competitive inhibition of a histone methyltransferase.
Chang, Y. et al. Adding a lysine mimic in the design of potent inhibitors of histone lysine methyltransferases. J. Mol. Biol. 400, 1–7 (2010).
Liu, F. et al. Protein lysine methyltransferase G9a inhibitors: design, synthesis, and structure activity relationships of 2,4-diamino-7-aminoalkoxy-quinazolines. J. Med. Chem. 53, 5844–5857 (2010).
Vedadi, M. et al. A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nature Chem. Biol. 7, 566–574 (2011).
Chang, Y. et al. Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294. Nature Struct. Mol. Biol. 16, 312–317 (2009).
Ferguson, A. D. et al. Structural basis of substrate methylation and inhibition of SMYD2. Structure 19, 1262–1273 (2011).
Greiner, D., Bonaldi, T., Eskeland, R., Roemer, E. & Imhof, A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3–9. Nature Chem. Biol. 1, 143–145 (2005).
Couture, J. F., Hauk, G., Thompson, M. J., Blackburn, G. M. & Trievel, R. C. Catalytic roles for carbon-oxygen hydrogen bonding in SET domain lysine methyltransferases. J. Biol. Chem. 281, 19280–19287 (2006).
Campagna-Slater, V. et al. Structural chemistry of the histone methyltransferases cofactor binding site. J. Chem. Inf. Model 51, 612–623 (2011).
Sack, J. S. et al. Structural basis for CARM1 inhibition by indole and pyrazole inhibitors. Biochem. J. 436, 331–339 (2011).
Dowden, J., Hong, W., Parry, R. V., Pike, R. A. & Ward, S. G. Toward the development of potent and selective bisubstrate inhibitors of protein arginine methyltransferases. Bioorg. Med. Chem. Lett. 20, 2103–2105 (2010).
Cheng, D. et al. Small molecule regulators of protein arginine methyltransferases. J. Biol. Chem. 279, 23892–23899 (2004).
Culhane, J. C., Wang, D., Yen, P. M. & Cole, P. A. Comparative analysis of small molecules and histone substrate analogues as LSD1 lysine demethylase inhibitors. J. Am. Chem. Soc. 132, 3164–3176 (2010).
Schmidt, D. M. & McCafferty, D. G. trans-2-Phenylcyclopropylamine is a mechanism-based inactivator of the histone demethylase LSD1. Biochemistry 46, 4408–4416 (2007).
Mimasu, S. et al. Structurally designed trans-2-phenylcyclopropylamine derivatives potently inhibit histone demethylase LSD1/KDM1. Biochemistry 49, 6494–6503 (2010).
Binda, C. et al. Biochemical, structural, and biological evaluation of tranylcypromine derivatives as inhibitors of histone demethylases LSD1 and LSD2. J. Am. Chem. Soc. 132, 6827–6833 (2010).
Ogasawara, D. et al. Synthesis and biological activity of optically active NCL-1, a lysine-specific demethylase 1 selective inhibitor. Bioorg. Med. Chem. 19, 3702–3708 (2011).
Ortega Muñoz, A., Castro- Palomino-Laria, J. & Fyfe, M. C. T. Lysine specific demethylase-1 inhibitors and their use. Patent WO2011035941A1 (2011).
Rose, N. R. et al. Inhibitor scaffolds for 2-oxoglutarate-dependent histone lysine demethylases. J. Med. Chem. 51, 7053–7056 (2008).
Luo, X. et al. A selective inhibitor and probe of the cellular functions of jumonji C domain-containing histone demethylases. J. Am. Chem. Soc. 133, 9451–9456 (2011).
Chang, K. H. et al. Inhibition of histone demethylases by 4-carboxy-2,2′-bipyridyl compounds. ChemMedChem 6, 759–764 (2011).
King, O. N. et al. Quantitative high-throughput screening identifies 8-hydroxyquinolines as cell-active histone demethylase inhibitors. PLoS ONE 5, e15535 (2010).
Ruthenburg, A. J., Li, H., Patel, D. J. & Allis, C. D. Multivalent engagement of chromatin modifications by linked binding modules. Nature Rev. Mol. Cell Biol. 8, 983–994 (2007).
Taverna, S. D., Li, H., Ruthenburg, A. J., Allis, C. D. & Patel, D. J. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nature Struct. Mol. Biol. 14, 1025–1040 (2007).
Jacobson, R. H., Ladurner, A. G., King, D. S. & Tjian, R. Structure and function of a human TAFII250 double bromodomain module. Science 288, 1422–1425 (2000).
Zeng, L. et al. Selective small molecules blocking HIV-1 Tat and coactivator PCAF association. J. Am. Chem. Soc. 127, 2376–2377 (2005).
Borah, J. C. et al. A small molecule binding to the coactivator CREB-binding protein blocks apoptosis in cardiomyocytes. Chem. Biol. 18, 531–541 (2011).
Chung, C. W. et al. Discovery and characterization of small molecule inhibitors of the BET family bromodomains. J. Med. Chem. 54, 3827–3838 (2011).
Herold, J. M. et al. Small-molecule ligands of methyl-lysine binding proteins. J. Med. Chem. 54, 2504–2511 (2011). This was the first demonstration that methyl-lysine binding pockets can be antagonized using small molecules.
Best, J. D. & Carey, N. Epigenetic opportunities and challenges in cancer. Drug Discov. Today 15, 65–70 (2010).
Best, J. D. & Carey, N. Epigenetic therapies for non-oncology indications. Drug Discov. Today 15, 1008–1014 (2010).
Anway, M. D., Cupp, A. S., Uzumcu, M. & Skinner, M. K. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 308, 1466–1469 (2005).
Anway, M. D., Leathers, C. & Skinner, M. K. Endocrine disruptor vinclozolin induced epigenetic transgenerational adult-onset disease. Endocrinology 147, 5515–5523 (2006).
Bertram, C. et al. Transgenerational effects of prenatal nutrient restriction on cardiovascular and hypothalamic-pituitary-adrenal function. J. Physiol. 586, 2217–2229 (2008).
Brower, V. Epigenetics: unravelling the cancer code. Nature 471, S12–S13 (2011).
Blanco, D. et al. Molecular analysis of a multistep lung cancer model induced by chronic inflammation reveals epigenetic regulation of p16 and activation of the DNA damage response pathway. Neoplasia 9, 840–852 (2007).
Richon, V. M. et al. Chemogenetic analysis of human protein methyltransferases. Chem. Biol. Drug Des. 78, 199–210 (2011).
Bamborough, P. et al. Fragment-based discovery of bromodomain inhibitors part 2: optimization of phenylisoxazole sulfonamides. J. Med. Chem. 55, 587–596 (2012).
Medda, F. et al. Novel cambinol analogs as sirtuin inhibitors: synthesis, biological evaluation, and rationalization of activity. J. Med. Chem. 52, 2673–2682 (2009).
Marks, P. A. & Breslow, R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nature Biotech. 25, 84–90 (2007).
Bertino, E. M. & Otterson, G. A. Romidepsin: a novel histone deacetylase inhibitor for cancer. Expert Opin. Investig. Drugs 20, 1151–1158 (2011).
Zhou, Q., Atadja, P. & Davidson, N. E. Histone deacetylase inhibitor LBH589 reactivates silenced estrogen receptor α (ER) gene expression without loss of DNA hypermethylation. Cancer Biol. Ther. 6, 64–69 (2007).
Plumb, J. A. et al. Pharmacodynamic response and inhibition of growth of human tumor xenografts by the novel histone deacetylase inhibitor PXD101. Mol. Cancer Ther. 2, 721–728 (2003).
Hu, E. et al. Identification of novel isoform-selective inhibitors within class I histone deacetylases. J. Pharmacol. Exp. Ther. 307, 720–728 (2003).
Fournel, M. et al. MGCD0103, a novel isotype-selective histone deacetylase inhibitor, has broad spectrum antitumor activity in vitro and in vivo. Mol. Cancer Ther. 7, 759–768 (2008).
Younes, A. et al. Mocetinostat for relapsed classical Hodgkin's lymphoma: an open-label, single-arm, Phase 2 trial. Lancet Oncol. 12, 1222–1228 (2011).
Mandl-Weber, S. et al. The novel inhibitor of histone deacetylase resminostat (RAS2410) inhibits proliferation and induces apoptosis in multiple myeloma (MM) cells. Br. J. Haematol. 149, 518–528 (2010).
Furlan, A. et al. Pharmacokinetics, safety and inducible cytokine responses during a Phase 1 trial of the oral histone deacetylase inhibitor ITF2357 (givinostat). Mol. Med. 17, 353–362 (2011).
Wang, H. et al. Discovery of (2E)-3-{2-butyl-1-[2-(diethylamino)ethyl]-1H-benzimidazol-5-yl}-N-hydroxya crylamide (SB939), an orally active histone deacetylase inhibitor with a superior preclinical profile. J. Med. Chem. 54, 4694–4720 (2011).
Novotny-Diermayr, V. et al. SB939, a novel potent and orally active histone deacetylase inhibitor with high tumor exposure and efficacy in mouse models of colorectal cancer. Mol. Cancer Ther. 9, 642–652 (2010).
Lai, C. J. et al. CUDC-101, a multitargeted inhibitor of histone deacetylase, epidermal growth factor receptor, and human epidermal growth factor receptor 2, exerts potent anticancer activity. Cancer Res. 70, 3647–3656 (2010).
Rivera-Del Valle, N. et al. PCI-24781, a novel hydroxamic acid HDAC inhibitor, exerts cytotoxicity and histone alterations via caspase-8 and FADD in leukemia cells. Int. J. Cell Biol. 2010, 207420 (2010).
Lucas, D. M. et al. The novel deacetylase inhibitor AR-42 demonstrates pre-clinical activity in B-cell malignancies in vitro and in vivo. PLoS ONE 5, e10941 (2010).
Hwang, J. J. et al. A novel histone deacetylase inhibitor, CG200745, potentiates anticancer effect of docetaxel in prostate cancer via decreasing Mcl-1 and Bcl-(XL). Invest. New Drugs 20 Jul 2011 (doi: 10.1007/s10637-011-9718-1).
Rosato, R. R. HDAC inhibitors — CHI's third annual conference. IDrugs 13, 13–15 (2010).
Acknowledgements
We are grateful to P. Brennan for his contribution on demethylase inhibitors, and S. Knapp for his comments on the manuscript. The Structural Genomics Consortium is a registered charity (charity number 1097737) that receives funds from the Canadian Institutes of Health Research, Eli Lilly, Genome Canada (through the Ontario Genomics Institute), GlaxoSmithKline, the Ontario Ministry for Research and Innovation, the Novartis Research Foundation, Pfizer and the Wellcome Trust.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
K.L. was an employee of GlaxoSmithKline and is now an employee of Pfizer. All other authors declare no competing financial interests.
Supplementary information
Supplementary information
BROMO_domain_fullname (XLS 150 kb)
Supplementary information
CHROMO_domain_fullname (XLS 95 kb)
Supplementary information
HAT_full_length_fullname (XLS 61 kb)
Supplementary information
HDAC_SIRT_domain_fullname (XLS 71 kb)
Supplementary information
KDM_domain_fullname (XLS 93 kb)
Supplementary information
MBT_domain_fullname (XLS 60 kb)
Supplementary information
PHD_domain_fullname (XLS 271 kb)
Supplementary information
PMT_domain_fullname (XLS 164 kb)
Supplementary information
PWWP_domain_fullname (XLS 74 kb)
Supplementary information
TUDOR_domain_fullname (XLS 119 kb)
Supplementary information
In format provided by Fish et al. (MAY 2012) (PDF 186 kb)
Supplementary information
In format provided by Fish et al. (MAY 2012) (PDF 555 kb)
Supplementary information
In format provided by Fish et al. (MAY 2012) (PDF 148 kb)
Supplementary information
In format provided by Fish et al. (MAY 2012) (PDF 211 kb)
Related links
Related links
FURTHER INFORMATION
EnVivo Pharmaceuticals website — Pipeline & Programs
RCSB Protein Data Bank website
RepliGen website (RG2833: Potential to Alter the Course of Friedreich's Ataxia)
Siena Biotech website — Development Pipeline
SMART (Simple Modular Architecture Research Tool) database
Structural Genomics Consortium (Annotated Phylogenetic Trees)
Structural Genomics Consortium (Chemical Probes)
Structural Genomics Consortium (Epigenetics: Chemical Probes for Drug Discovery)
Glossary
- Chromatin
-
The fibres in which DNA and genes are packaged in the nucleus of a cell. Chromatin consists of the DNA double helix wrapped around a complex of histone proteins — together called the nucleosome.
- Epigenetics
-
Heritable changes in gene expression or phenotype that are stable between cell divisions, and sometimes between generations, but do not involve changes in the underlying DNA sequence of the organism.
- Differentiation
-
The process by whicha stem cell, or other precursor cell, commits towards a more specialized cell type with a specific function, and represents an exit from self-renewal. Differentiation is controlled by cell signalling pathways and maintained through epigenetic mechanisms.
- Post-translational modification
-
A chemical modification of proteins that acts as a signal to other proteins that recognize the modification. In the context of epigenetic signalling, post-translational modifications are often called 'marks'.
- Epigenome
-
The combination of histone and DNA post-translational modifications and related interacting proteins that together package the genome and help define the transcriptional programme in a given cell.
- Heterochromatin
-
A tightly packed form of DNA associated with transcriptionally silent or repressed genes. It is highly correlated with di- and trimethlyated H3K9 (Lys9 of histone 3) marks.
- Euchromatin
-
A more loosely packed form of DNA that is associated with transcriptionally active genes.
- Bromodomain
-
An evolutionarily conserved, ∼110-amino-acid motif composed of four left-handed, antiparallel α-helices.
- Stem cell
-
An unspecialized precursor cell withthe capacity to self-renew (continuously produce unaltered progeny) and to differentiate into more mature specialized cell types.
- Haploinsufficiency
-
A disease mechanism in which one of two copies of a gene is mutated, resulting in insufficient activity of the gene products (typically a protein) to bring about a functional, wild-type condition.
- Brachydactyly mental retardation syndrome
-
A disorder that presents with a range of features, including intellectual disabilities, developmental delays, behavioural abnormalities, sleep disturbance, craniofacial and skeletal abnormalities, and autism spectrum disorder.
- Presenilins
-
A family of related multipass transmembrane proteins that function as part of the γ-secretase intramembrane protease complex. They were first identified in screens for mutations causing early-onset forms of familial Alzheimer's disease.
- Amyotrophic lateral sclerosis
-
A progressive neurological disease that is associated with the degeneration of central and spinal motor neurons. This neuron loss causes muscles to weaken and waste away, leading to paralysis.
- Jumonji domain
-
A conserved domain originally identified in the Jumonji family of transcription factors, now known to be histone demethylases. The Jumonji C domain comprises the catalytic domain of the 2-oxoglutarate-dependent lysine demethylases.
- Hyperglycaemic memory
-
A phenomenon in which the deleterious end-organ effects resulting from exposure to high glucose levels persist for several years after usual glycaemic control is restored.
- Malignant brain tumour domains
-
Conserved sequence motifs found in certain developmental proteins. These domains bind to mono- or dimethylated lysine-containing peptides and, when deleted in fruitflies, lead to brain tumours.
- Allosteric stimulation
-
The regulation of an enzyme or protein by binding an effector molecule at a site other than the protein's active site, thereby causing a conformational change in the protein.
- P300/CBP-associated factor
-
A trancriptional co-activator protein containing a bromodomain and a histone acetyltransferase domain.
- π electron interactions
-
A non-covalent interaction between the π-electron cloud of aromatic rings and the cationic charge of, for example, methylated lysine.
Rights and permissions
About this article
Cite this article
Arrowsmith, C., Bountra, C., Fish, P. et al. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov 11, 384–400 (2012). https://doi.org/10.1038/nrd3674
Published:
Issue date:
DOI: https://doi.org/10.1038/nrd3674
This article is cited by
-
3D Models of Sarcomas: The Next-generation Tool for Personalized Medicine
Phenomics (2024)
-
Combined low levels of H4K16ac and H4K20me3 predicts poor prognosis in breast cancer
International Journal of Clinical Oncology (2023)
-
Structural classification of EZH2 inhibitors and prospects for the treatment of tumor: a review
Medicinal Chemistry Research (2023)
-
The role of histone methylation in renal cell cancer: an update
Molecular Biology Reports (2023)
-
Fabrication and characterization of bilayer scaffolds made of decellularized dermis/nanofibrous collagen for healing of full-thickness wounds
Drug Delivery and Translational Research (2023)