Key Points
-
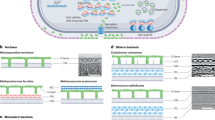
S-layers are two-dimensional (2D) protein arrays that are frequently found on the surface of bacteria and archaea.
-
Genetic analysis reveals a wide diversity of genes that encode S-layer proteins (SLPs) in some species, and several mechanisms are found to facilitate gene switching and regulation.
-
Secretion of S-layer proteins often involves a dedicated secretion system, such as accessory Sec systems in Bacillus anthracis and Clostridium difficile, and a wide range of mechanisms for anchoring S-layers to the underlying cell envelope have been identified.
-
Gram-positive species, including B. anthracis and C. difficile, possess large families of genes encoding proteins that are related to the S-layer protein and that share a common anchoring mechanism.
-
In many species, the SLPs are glycosylated. Dedicated glycosylation loci are found that specify all the genes that are necessary for the synthesis of glycan, its secretion across the membrane and ligation to the SLP via N- or O- linkages.
-
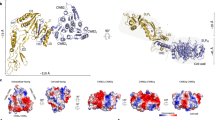
S-layers have been the subject of intensive structural analysis since their identification in the 1950s. Recent efforts are gradually improving our high-resolution structural knowledge of various S-layer proteins and finally enabling reasonable quality models of an entire S-layer to be made.
-
SLPs have evolved to mediate a broad range of functions, including biogenesis of the cell wall, control of cell division and specialized activities, such as swimming. In pathogens, SLPs can interfere with the immune system and can aid survival via adhesion to host cells. In Gram-positive bacteria, functions are often associated with an effector domain that can confer properties that are distinct from the ability to form a 2D array.
Abstract
The outer surface of many archaea and bacteria is coated with a proteinaceous surface layer (known as an S-layer), which is formed by the self-assembly of monomeric proteins into a regularly spaced, two-dimensional array. Bacteria possess dedicated pathways for the secretion and anchoring of the S-layer to the cell wall, and some Gram-positive species have large S-layer-associated gene families. S-layers have important roles in growth and survival, and their many functions include the maintenance of cell integrity, enzyme display and, in pathogens and commensals, interaction with the host and its immune system. In this Review, we discuss our current knowledge of S-layer and related proteins, including their structures, mechanisms of secretion and anchoring and their diverse functions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Sara, M. & Sleytr, U. B. S-layer proteins. J. Bacteriol. 182, 859–868 (2000).
Sleytr, U. B. & Beveridge, T. J. Bacterial S-layers. Trends Microbiol. 7, 253–260 (1999).
Albers, S. V. & Meyer, B. H. The archaeal cell envelope. Nature Rev. Microbiol. 9, 414–426 (2011).
Sleytr, U. B. & Glauert, A. M. Ultrastructure of the cell walls of two closely related Clostridia that possess different regular arrays of surface subunits. J. Bacteriol. 126, 869–882 (1976).
Sleytr, U. B. et al. S-layers as a tool kit for nanobiotechnological applications. FEMS Microbiol. Lett. 267, 131–144 (2007).
Schuster, B. & Sleytr, U. B. Nanotechnology with S-layer proteins. Methods Mol. Biol. 996, 153–175 (2013).
Tu, Z. C., Wassenaar, T. M., Thompson, S. A. & Blaser, M. J. Structure and genotypic plasticity of the Campylobacter fetus sap locus. Mol. Microbiol. 48, 685–698 (2003).
Dworkin, J. & Blaser, M. J. Nested DNA inversion as a paradigm of programmed gene rearrangement. Proc. Natl Acad. Sci. USA 94, 985–990 (1997).
Tummuru, M. K. & Blaser, M. J. Rearrangement of sapA homologs with conserved and variable regions in Campylobacter fetus. Proc. Natl Acad. Sci. USA 90, 7265–7269 (1993). This paper provides the first description of site-specific recombination between SLP genes as a mechanism for antigenic variation.
Dworkin, J., Tummuru, M. K. & Blaser, M. J. Segmental conservation of sapA sequences in type B Campylobacter fetus cells. J. Biol. Chem. 270, 15093–15101 (1995).
Eidhin, D., Ryan, A., Doyle, R., Walsh, J. B. & Kelleher, D. Sequence and phylogenetic analysis of the gene for surface layer protein, slpA, from 14 PCR ribotypes of Clostridium difficile. J. Med. Microbiol. 55, 69–83 (2006).
Calabi, E. & Fairweather, N. Patterns of sequence conservation in the S-layer proteins and related sequences in Clostridium difficile. J. Bacteriol. 184, 3886–3897 (2002).
Dingle, K. E. et al. Recombinational switching of the Clostridium difficile S-layer and a novel glycosylation gene cluster revealed by large-scale whole-genome sequencing. J. Infect. Dis. 207, 675–686 (2013). This paper provides a description of SLP cassettes in C. difficile , with genetic evidence of recombinational switching, which is hypothesized to facilitate antigenic variation.
Emerson, J. et al. A novel genetic switch controls phase variable expression of CwpV, a Clostridium difficile cell wall protein. Mol. Microbiol. 74, 541–556 (2009).
Reynolds, C. B., Emerson, J. E., de la Riva, L., Fagan, R. P. & Fairweather, N. F. The Clostridium difficile cell wall protein CwpV is antigenically variable between strains, but exhibits conserved aggregation-promoting function. PLoS Pathog. 7, e1002024 (2011).
Kern, J. W. & Schneewind, O. BslA, a pXO1-encoded adhesin of Bacillus anthracis. Mol. Microbiol. 68, 504–515 (2008).
Mignot, T., Mesnage, S., Couture-Tosi, E., Mock, M. & Fouet, A. Developmental switch of S-layer protein synthesis in Bacillus anthracis. Mol. Microbiol. 43, 1615–1627 (2002).
Wang, Y. T., Oh, S. Y., Hendrickx, A. P., Lunderberg, J. M. & Schneewind, O. Bacillus cereus G9241 S-layer assembly contributes to the pathogenesis of anthrax-like disease in mice. J. Bacteriol. 195, 596–605 (2012).
Fagan, R. P. et al. A proposed nomenclature for cell wall proteins of Clostridium difficile. J. Med. Microbiol. 60, 1225–1228 (2011).
Bruggemann, H. et al. The genome sequence of Clostridium tetani, the causative agent of tetanus disease. Proc. Natl Acad. Sci. USA 100, 1316–1321 (2003).
Sebaihia, M. et al. Genome sequence of a proteolytic (Group I) Clostridium botulinum strain Hall A and comparative analysis of the clostridial genomes. Genome Res. 17, 1082–1092 (2007).
Awram, P. & Smit, J. The Caulobacter crescentus paracrystalline S-layer protein is secreted by an ABC transporter (type I) secretion apparatus. J. Bacteriol. 180, 3062–3069 (1998).
Kawai, E., Akatsuka, H., Idei, A., Shibatani, T. & Omori, K. Serratia marcescens S-layer protein is secreted extracellularly via an ATP-binding cassette exporter, the Lip system. Mol. Microbiol. 27, 941–952 (1998).
Thompson, S. A. et al. Campylobacter fetus surface layer proteins are transported by a type I secretion system. J. Bacteriol. 180, 6450–6458 (1998).
Noonan, B. & Trust, T. J. Molecular analysis of an A-protein secretion mutant of Aeromonas salmonicida reveals a surface layer-specific protein secretion pathway. J. Mol. Biol. 248, 316–327 (1995).
Thomas, S. R. & Trust, T. J. A specific PulD homolog is required for the secretion of paracrystalline surface array subunits in Aeromonas hydrophila. J. Bacteriol. 177, 3932–3939 (1995).
Nguyen-Mau, S. M., Oh, S. Y., Kern, V. J., Missiakas, D. M. & Schneewind, O. Secretion genes as determinants of Bacillus anthracis chain length. J. Bacteriol. 194, 3841–3850 (2012).
Fagan, R. P. & Fairweather, N. F. Clostridium difficile has two parallel and essential Sec secretion systems. J. Biol. Chem. 286, 27483–27493 (2011).
Braunstein, M., Brown, A. M., Kurtz, S. & Jacobs, W. R. Jr. Two nonredundant SecA homologues function in Mycobacteria. J. Bacteriol. 183, 6979–6990 (2001).
Feltcher, M. E. & Braunstein, M. Emerging themes in SecA2-mediated protein export. Nature Rev. Microbiol. 10, 779–789 (2012).
Awram, P. & Smit, J. Identification of lipopolysaccharide O antigen synthesis genes required for attachment of the S-layer of Caulobacter crescentus. Microbiology 147, 1451–1460 (2001).
Ford, M. J., Nomellini, J. F. & Smit, J. S-layer anchoring and localization of an S-layer-associated protease in Caulobacter crescentus. J. Bacteriol. 189, 2226–2237 (2007).
Yang, L. Y., Pei, Z. H., Fujimoto, S. & Blaser, M. J. Reattachment of surface array proteins to Campylobacter fetus cells. J. Bacteriol. 174, 1258–1267 (1992).
Dworkin, J., Tummuru, M. K. & Blaser, M. J. A lipopolysaccharide-binding domain of the Campylobacter fetus S-layer protein resides within the conserved N terminus of a family of silent and divergent homologs. J. Bacteriol. 177, 1734–1741 (1995).
Ebisu, S. et al. Conserved structures of cell wall protein genes among protein-producing Bacillus brevis strains. J. Bacteriol. 172, 1312–1320 (1990).
Bowditch, R. D., Baumann, P. & Yousten, A. A. Cloning and sequencing of the gene encoding a 125-kilodalton surface-layer protein from Bacillus sphaericus 2362 and of a related cryptic gene. J. Bacteriol. 171, 4178–4188 (1989).
Faraldo, M. M., de Pedro, M. A. & Berenguer, J. Sequence of the S-layer gene of Thermus thermophilus HB8 and functionality of its promoter in Escherichia coli. J. Bacteriol. 174, 7458–7462 (1992).
Kuen, B., Sleytr, U. B. & Lubitz, W. Sequence analysis of the sbsA gene encoding the 130-kDa surface-layer protein of Bacillus stearothermophilus strain PV72. Gene 145, 115–120 (1994).
Lemaire, M., Miras, I., Gounon, P. & Beguin, P. Identification of a region responsible for binding to the cell wall within the S-layer protein of Clostridium thermocellum. Microbiology 144, 211–217 (1998).
Mesnage, S. et al. Bacterial SLH domain proteins are non-covalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. EMBO J. 19, 4473–4484 (2000). This paper gives the first description of a cell wall ligand for an SLP — a pyruvylated SCWP as the ligand for non-covalent anchoring of B. anthracis SLPs that contain SLH domains.
Etienne-Toumelin, I., Sirard, J. C., Duflot, E., Mock, M. & Fouet, A. Characterization of the Bacillus anthracis S-layer: cloning and sequencing of the structural gene. J. Bacteriol. 177, 614–620 (1995).
Mesnage, S., Tosi-Couture, E., Mock, M., Gounon, P. & Fouet, A. Molecular characterization of the Bacillus anthracis main S-layer component: evidence that it is the major cell-associated antigen. Mol. Microbiol. 23, 1147–1155 (1997).
Kern, J. et al. Structure of the SLH domains from Bacillus anthracis surface array protein. J. Biol. Chem. 286, 26042–26049 (2011).
Sara, M. et al. Dynamics in oxygen-induced changes in S-layer protein synthesis from Bacillus stearothermophilus PV72 and the S-layer-deficient variant T5 in continuous culture and studies of the cell wall composition. J. Bacteriol. 178, 2108–2117 (1996).
Jarosch, M., Egelseer, E. M., Mattanovich, D., Sleytr, U. B. & Sara, M. S-layer gene sbsC of Bacillus stearothermophilus ATCC 12980: molecular characterization and heterologous expression in Escherichia coli. Microbiology 146, 273–281 (2000).
Egelseer, E. M. et al. Characterization of an S-layer glycoprotein produced in the course of S-layer variation of Bacillus stearothermophilus ATCC 12980 and sequencing and cloning of the sbsD gene encoding the protein moiety. Arch. Microbiol. 177, 70–80 (2001).
Schaffer, C. et al. The surface layer (S-layer) glycoprotein of Geobacillus stearothermophilus NRS 2004/3a. Analysis of its glycosylation. J. Biol. Chem. 277, 6230–6239 (2002).
Mader, C., Huber, C., Moll, D., Sleytr, U. B. & Sara, M. Interaction of the crystalline bacterial cell surface layer protein SbsB and the secondary cell wall polymer of Geobacillus stearothermophilus PV72 assessed by real-time surface plasmon resonance biosensor technology. J. Bacteriol. 186, 1758–1768 (2004).
Schaffer, C. et al. The diacetamidodideoxyuronic-acid-containing glycan chain of Bacillus stearothermophilus NRS 2004/3a represents the secondary cell-wall polymer of wild-type B. stearothermophilus strains. Microbiology 145, 1575–1583 (1999).
Ferner-Ortner, J., Mader, C., Ilk, N., Sleytr, U. B. & Egelseer, E. M. High-affinity interaction between the S-layer protein SbsC and the secondary cell wall polymer of Geobacillus stearothermophilus ATCC 12980 determined by surface plasmon resonance technology. J. Bacteriol. 189, 7154–7158 (2007).
Kuroda, A., Rashid, M. H. & Sekiguchi, J. Molecular cloning and sequencing of the upstream region of the major Bacillus subtilis autolysin gene: a modifier protein exhibiting sequence homology to the major autolysin and the spoIID product. J. Gen. Microbiol. 138, 1067–1076 (1992).
Kuroda, A. & Sekiguchi, J. Cloning, sequencing and genetic mapping of a Bacillus subtilis cell wall hydrolase gene. J. Gen. Microbiol. 136, 2209–2216 (1990).
Bruggemann, H. & Gottschalk, G. Comparative genomics of Clostridia: link between the ecological niche and cell surface properties. Ann. NY Acad. Sci. 1125, 73–81 (2008).
Weidenmaier, C. & Peschel, A. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nature Rev. Microbiol. 6, 276–287 (2008).
Pavkov, T. et al. The structure and binding behavior of the bacterial cell surface layer protein SbsC. Structure 16, 1226–1237 (2008).
Runzler, D., Huber, C., Moll, D., Kohler, G. & Sara, M. Biophysical characterization of the entire bacterial surface layer protein SbsB and its two distinct functional domains. J Biol Chem, 7, 5207–5215 (2003).
Mignot, T. et al. Distribution of S-layers on the surface of Bacillus cereus strains: phylogenetic origin and ecological pressure. Environ. Microbiol. 3, 493–501 (2001).
Baranova, E. et al. SbsB structure and lattice reconstruction unveil Ca2+ triggered S-layer assembly. Nature 487, 119–122 (2012). This paper provides a high-resolution model of the assembled G. stearothermophilus SbsB S-layer, extrapolated from X-ray crystallography. It also gives a plausible explanation for the calcium-dependence of SLP self-assembly.
Mescher, M. F., Strominger, J. L. & Watson, S. W. Protein and carbohydrate composition of the cell envelope of Halobacterium salinarium. J. Bacteriol. 120, 945–954 (1974).
Ristl, R. et al. The S-layer glycome — adding to the sugar coat of bacteria. Int J. Microbiol 2011, 127870 (2011). This study provides the first complete description of a pathway for glycosylation of an S-layer protein.
Messner, P., Steiner, K., Zarschler, K. & Schaffer, C. S-layer nanoglycobiology of bacteria. Carbohydr. Res. 343, 1934–1951 (2008).
Abu-Qarn, M., Eichler, J. & Sharon, N. Not just for eukarya anymore: protein glycosylation in bacteria and archaea. Curr. Opin. Struct. Biol. 18, 544–550 (2008).
Benz, I. & Schmidt, M. A. Never say never again: protein glycosylation in pathogenic bacteria. Mol. Microbiol. 45, 267–276 (2002).
Schaffer, C., Wugeditsch, T., Neuninger, C. & Messner, P. Are S-layer glycoproteins and lipopolysaccharides related? Microb. Drug Resist. 2, 17–23 (1996).
Steiner, K. et al. Molecular basis of S-layer glycoprotein glycan biosynthesis in Geobacillus stearothermophilus. J. Biol. Chem. 283, 21120–21133 (2008).
Zarschler, K., Janesch, B., Zayni, S., Schaffer, C. & Messner, P. Construction of a gene knockout system for application in Paenibacillus alvei CCM 2051T, exemplified by the S-layer glycan biosynthesis initiation enzyme WsfP. Appl. Environ. Microbiol. 75, 3077–3085 (2009).
Posch, G. et al. Characterization and scope of S-layer protein O-glycosylation in Tannerella forsythia. J. Biol. Chem. 286, 38714–38724 (2011).
Qazi, O. et al. Mass spectrometric analysis of the S-layer proteins from Clostridium difficile demonstrates the absence of glycosylation. J. Mass Spectrom. 44, 368–374 (2009).
Beveridge, T. J. et al. Functions of S-layers. FEMS Microbiol. Rev. 20, 99–149 (1997).
Sun, Z. et al. Characterization of a S-layer protein from Lactobacillus crispatus K313 and the domains responsible for binding to cell wall and adherence to collagen. Appl. Microbiol. Biotechnol. 97, 1941–1952 (2013).
Sillanpaa, J. et al. Characterization of the collagen-binding S-layer protein CbsA of Lactobacillus crispatus. J. Bacteriol. 182, 6440–6450 (2000).
Toba, T. et al. A collagen-binding S-layer protein in Lactobacillus crispatus. Appl. Environ. Microbiol. 61, 2467–2471 (1995).
Ausiello, C. M. et al. Surface layer proteins from Clostridium difficile induce inflammatory and regulatory cytokines in human monocytes and dendritic cells. Microbes Infect. 8, 2640–2646 (2006).
Ryan, A. et al. A role for TLR4 in Clostridium difficile infection and the recognition of surface layer proteins. PLoS Pathog 7, e1002076 (2011). This study shows that an SLP can function as a ligand for the innate immune response, activating TLR4 and promoting clearance of C. difficile infection.
Calabi, E., Calabi, F., Phillips, A. D. & Fairweather, N. Binding of Clostridium difficile surface layer proteins to gastrointestinal tissues. Infect. Immun. 70, 5770–5778 (2002).
Faulds-Pain, A. & Wren, B. W. Improved bacterial mutagenesis by high-frequency allele exchange, demonstrated in Clostridium difficile and Streptococcus suis. Appl. Environ. Microbiol. 79, 4768–4771 (2013).
Cartman, S. T., Kelly, M. L., Heeg, D., Heap, J. T. & Minton, N. P. Precise manipulation of the Clostridium difficile chromosome reveals a lack of association between tcdC genotype and toxin production. Appl. Environ. Microbiol. 78, 4683–4690 (2012).
Heap, J. T. et al. Integration of DNA into bacterial chromosomes from plasmids without a counter-selection marker. Nucleic Acids Res. 40, e59 (2012).
Kern, J. & Schneewind, O. BslA, the S-layer adhesin of B. anthracis, is a virulence factor for anthrax pathogenesis. Mol. Microbiol. 75, 324–332 (2010).
Tarlovsky, Y. et al. A Bacillus anthracis S-layer homology protein that binds heme and mediates heme delivery to IsdC. J. Bacteriol. 192, 3503–3511 (2010).
Grogono-Thomas, R., Dworkin, J., Blaser, M. J. & Newell, D. G. Roles of the surface layer proteins of Campylobacter fetus subsp. fetus in ovine abortion. Infect. Immun. 68, 1687–1691 (2000).
Garcia, M. M. et al. Protein shift and antigenic variation in the S-layer of Campylobacter fetus subsp. venerealis during bovine infection accompanied by genomic rearrangement of sapA homologs. J. Bacteriol. 177, 1976–1980 (1995).
Wang, E., Garcia, M. M., Blake, M. S., Pei, Z. & Blaser, M. J. Shift in S-layer protein expression responsible for antigenic variation in Campylobacter fetus. J. Bacteriol. 175, 4979–4984 (1993).
Blaser, M. J. & Pei, Z. Pathogenesis of Campylobacter fetus infections: critical role of high-molecular-weight S-layer proteins in virulence. J. Infect. Dis. 167, 372–377 (1993).
Blaser, M. J. et al. Pathogenesis of Campylobacter fetus infections: serum resistance associated with high-molecular-weight surface proteins. J. Infect. Dis. 155, 696–706 (1987).
Blaser, M. J., Smith, P. F., Repine, J. E. & Joiner, K. A. Pathogenesis of Campylobacter fetus infections. Failure of encapsulated Campylobacter fetus to bind C3b explains serum and phagocytosis resistance. J. Clin. Invest. 81, 1434–1444 (1988).
Tu, Z. C., Gaudreau, C. & Blaser, M. J. Mechanisms underlying Campylobacter fetus pathogenesis in humans: surface-layer protein variation in relapsing infections. J. Infect. Dis. 191, 2082–2089 (2005).
Darveau, R. P. Periodontitis: a polymicrobial disruption of host homeostasis. Nature Rev. Microbiol. 8, 481–490 (2010).
Socransky, S. S., Haffajee, A. D., Cugini, M. A., Smith, C. & Kent, R. L. Jr. Microbial complexes in subgingival plaque. J. Clin. Periodontol 25, 134–144 (1998).
Sabet, M., Lee, S. W., Nauman, R. K., Sims, T. & Um, H. S. The surface (S-) layer is a virulence factor of Bacteroides forsythus. Microbiology 149, 3617–3627 (2003).
Higuchi, N. et al. Localization of major, high molecular weight proteins in Bacteroides forsythus. Microbiol. Immunol. 44, 777–780 (2000).
Sakakibara, J. et al. Loss of adherence ability to human gingival epithelial cells in S-layer protein-deficient mutants of Tannerella forsythensis. Microbiology 153, 866–876 (2007).
Lee, S. W. et al. Identification and characterization of the genes encoding a unique surface (S-) layer of Tannerella forsythia. Gene 371, 102–111 (2006).
Honma, K., Inagaki, S., Okuda, K., Kuramitsu, H. K. & Sharma, A. Role of a Tannerella forsythia exopolysaccharide synthesis operon in biofilm development. Microb. Pathog. 42, 156–166 (2007).
Pham, T. K. et al. A quantitative proteomic analysis of biofilm adaptation by the periodontal pathogen Tannerella forsythia. Proteomics 10, 3130–3141 (2010).
Sockett, R. E. Predatory lifestyle of Bdellovibrio bacteriovorus. Annu. Rev. Microbiol. 63, 523–539 (2009).
Koval, S. F. & Hynes, S. H. Effect of paracrystalline protein surface layers on predation by Bdellovibrio bacteriovorus. J. Bacteriol. 173, 2244–2249 (1991).
Sara, M. & Sleytr, U. B. Molecular sieving through S layers of Bacillus stearothermophilus strains. J. Bacteriol. 169, 4092–4098 (1987).
Rothfuss, H., Lara, J. C., Schmid, A. K. & Lidstrom, M. E. Involvement of the S-layer proteins Hpi and SlpA in the maintenance of cell envelope integrity in Deinococcus radiodurans R1. Microbiology 152, 2779–2787 (2006).
Kirby, J. M. et al. Cwp84, a surface-associated cysteine protease, plays a role in the maturation of the surface layer of Clostridium difficile. J. Biol. Chem. 284, 34666–34673 (2009).
de la Riva, L., Willing, S. E., Tate, E. W. & Fairweather, N. F. Roles of cysteine proteases Cwp84 and Cwp13 in biogenesis of the cell wall of Clostridium difficile. J. Bacteriol. 193, 3276–3285 (2011).
Ahn, J. S., Chandramohan, L., Liou, L. E. & Bayles, K. W. Characterization of CidR-mediated regulation in Bacillus anthracis reveals a previously undetected role of S-layer proteins as murein hydrolases. Mol. Microbiol. 62, 1158–1169 (2006).
Anderson, V. J., Kern, J. W., McCool, J. W., Schneewind, O. & Missiakas, D. The SLH-domain protein BslO is a determinant of Bacillus anthracis chain length. Mol. Microbiol. 81, 192–205 (2011).
Kern, V. J., Kern, J. W., Theriot, J. A., Schneewind, O. & Missiakas, D. Surface-layer (S-layer) proteins sap and EA1 govern the binding of the S-layer-associated protein BslO at the cell septa of Bacillus anthracis. J. Bacteriol. 194, 3833–3840 (2012).
Peltier, J. et al. Clostridium difficile has an original peptidoglycan structure with a high level of N-acetylglucosamine deacetylation and mainly 3–3 cross-links. J. Biol. Chem. 286, 29053–29062 (2011).
Brahamsha, B. An abundant cell-surface polypeptide is required for swimming by the nonflagellated marine cyanobacterium Synechococcus. Proc. Natl Acad. Sci. USA 93, 6504–6509 (1996).
McCarren, J. & Brahamsha, B. SwmB, a 1.12-megadalton protein that is required for nonflagellar swimming motility in Synechococcus. J. Bacteriol. 189, 1158–1162 (2007).
McCarren, J. & Brahamsha, B. Swimming motility mutants of marine Synechococcus affected in production and localization of the S-layer protein SwmA. J. Bacteriol. 191, 1111–1114 (2009).
Norville, J. E. et al. 7Å projection map of the S-layer protein sbpA obtained with trehalose-embedded monolayer crystals. J. Struct. Biol. 160, 313–323 (2007).
Houwink, A. L. A macromolecular mono-layer in the cell wall of Spirillum spec. Biochim. Biophys. Acta 10, 360–366 (1953). This paper provides the first description of a paracrystalline layer on a bacterial cell.
Severs, N. J. Freeze-fracture electron microscopy. Nature Protoc. 2, 547–576 (2007).
Sleytr, U. B., Messner, P., Pum, D. & Sara, M. Crystalline bacterial cell surface layers (S Layers): from supramolecular cell structure to biomimetics and nanotechnology. Angew. Chem. Int. Ed. 38, 1034–1054 (1999).
Lupas, A. et al. Domain structure of the Acetogenium kivui surface-layer revealed by electron crystallography and sequence-analysis. J. Bacteriol. 176, 1224–1233 (1994).
Dorobantu, L. S., Goss, G. G. & Burrell, R. E. Atomic force microscopy: a nanoscopic view of microbial cell surfaces. Micron 43, 1312–1322 (2012).
Chung, S., Shin, S. H., Bertozzi, C. R. & De Yoreo, J. J. Self-catalyzed growth of S layers via an amorphous-to-crystalline transition limited by folding kinetics. Proc. Natl Acad. Sci. USA 107, 16536–16541 (2010).
Stetefeld, J. et al. Crystal structure of a naturally occurring parallel right-handed coiled coil tetramer. Nature Struct. Biol. 7, 772–776 (2000).
Jing, H. et al. Archaeal surface layer proteins contain β propeller, PKD, and β helix domains and are related to metazoan cell surface proteins. Structure 10, 1453–1464 (2002).
Arbing, M. A. et al. Structure of the surface layer of the methanogenic archaean Methanosarcina acetivorans. Proc. Natl Acad. Sci. USA 109, 11812–11817 (2012).
Fagan, R. P. et al. Structural insights into the molecular organization of the S-layer from Clostridium difficile. Mol. Microbiol. 71, 1308–1322 (2009).
Punta, M. et al. The Pfam protein families database. Nucleic Acids Res. 40, D290–D301 (2012).
Korotkov, K. V., Sandkvist, M. & Hol, W. G. The type II secretion system: biogenesis, molecular architecture and mechanism. Nature Rev. Microbiol. 10, 336–351 (2012).
Acknowledgements
Work in the N.F.F. laboratory is currently supported by the MRC (grants G0800170 and G1001721), the Wellcome Trust (grant 090969Z) and the Leverhulme Trust (grant RF2012-232), and work in the R.P.F. laboratory is supported by the University of Sheffield, UK.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information S1 (table)
Identification of putative functional domains in the B. anthracis and C. difficile cell surface protein families (PDF 206 kb)
Glossary
- Phase-variable expression
-
The random variation of gene expression in a bacterial population. Expression in individual cells is either on or off, leading to phenotypic heterogeneity in the population.
- Type I secretion system
-
A sec-independent protein secretion system in Gram-negative bacteria. It consists of an inner membrane ATP-binding cassette (ABC) transporter, a periplasmic membrane fusion protein and an outer membrane pore.
- Type II secretion systems
-
sec-dependent multiprotein secretion systems in Gram-negative bacteria; they are closely related to type IV pili.
- Secondary cell wall polymer
-
(SCWP). A carbohydrate-based polymer, other than peptidoglycan and anionic polymers, that is present in the cell wall — for example, the pyruvylated Bacillus anthracis SCWP that anchors the S-layer proteins EA1 and Sap to the cell wall.
- N- and O-linkages
-
Linkage of a sugar to the nitrogen (N) atom of asparagine or to the oxygen (O) atom of serine, threonine or tyrosine.
- Sacculi
-
The sacs of polymerized peptidoglycan that surround bacteria. When they are isolated from the bacterium, they retain the shape of the cell.
Rights and permissions
About this article
Cite this article
Fagan, R., Fairweather, N. Biogenesis and functions of bacterial S-layers. Nat Rev Microbiol 12, 211–222 (2014). https://doi.org/10.1038/nrmicro3213
Published:
Issue date:
DOI: https://doi.org/10.1038/nrmicro3213
This article is cited by
-
Prokaryotic microvesicles Ortholog of eukaryotic extracellular vesicles in biomedical fields
Cell Communication and Signaling (2024)
-
Shape control in 2D molecular nanosheets by tuning anisotropic intermolecular interactions and assembly kinetics
Nature Communications (2023)
-
Previously uncharacterized rectangular bacterial structures in the dolphin mouth
Nature Communications (2023)
-
Role of cell-substrate association during plant biomass solubilization by the extreme thermophile Caldicellulosiruptor bescii
Extremophiles (2023)
-
Structure and assembly of the S-layer in C. difficile
Nature Communications (2022)