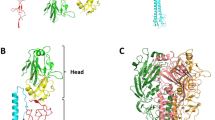
Key Points
-
Interspecies transmission of influenza A viruses is the result of many factors. One of the key factors involved is a shift in the receptor-binding specificity of the virus, which is mostly determined by mutations in viral haemagglutinin (HA).
-
Recent structural studies have provided molecular insights into the HA–host receptor interactions that have enabled several influenza A virus subtypes to jump from avian to human hosts.
-
The combination of distinct amino acids at positions 225 and 190 of HA is important for determining the receptor-binding specificity of the H1 subtype.
-
The Q226L and G228S substitutions in the HA glycoproteins of the H2 and H3 subtypes are sufficient to change the binding preference from the avian receptor to the human receptor.
-
In an experimentally adapted H5 subtype, the Q226L substitution and loss of a glycosylation site near the receptor-binding site contribute to the shift in binding preference from the avian receptor to the human receptor.
-
In the H7 subtype, amino acid substitutions at positions 186 and 226 of HA increase the preference for binding to the human receptor.
Abstract
The recent emergence of the H7N9 avian influenza A virus and its ability to infect humans emphasize the epidemic and pandemic potential of these viruses. Interspecies transmission is the result of many factors, which ultimately lead to a change in the host tropism of the virus. One of the key factors involved is a shift in the receptor-binding specificity of the virus, which is mostly determined by mutations in the viral haemagglutinin (HA). In this Review, we discuss recent crystallographic studies that provide molecular insights into HA–host receptor interactions that have enabled several influenza A virus subtypes to 'jump' from avian to human hosts.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Taubenberger, J. K. & Morens, D. M. Pandemic influenza — including a risk assessment of H5N1. Rev. Sci. Tech. 28, 187–202 (2009).
Wu, Y. & Gao, G. F. Lessons learnt from the human infections of avian-origin influenza A H7N9 virus: live free markets and human health. Sci. China Life Sci. 56, 493–494 (2013).
Liu, X., Zhao, Z. & Liu, W. Insights into the roles of cyclophilin A during influenza virus infection. Viruses 5, 182–191 (2013).
Wu, Y., Wu, Y., Tefsen, B., Shi, Y. & Gao, G. F. Bat-derived influenza-like viruses H17N10 and H18N11. Trends Microbiol. 22, 183–191 (2014).
Muramoto, Y., Noda, T., Kawakami, E., Akkina, R. & Kawaoka, Y. Identification of novel influenza A virus proteins translated from PA mRNA. J. Virol. 87, 2455–2462 (2013).
Gamblin, S. J. & Skehel, J. J. Influenza hemagglutinin and neuraminidase membrane glycoproteins. J. Biol. Chem. 285, 28403–28409 (2010).
Tong, S. et al. A distinct lineage of influenza A virus from bats. Proc. Natl Acad. Sci. USA 109, 4269–4274 (2012).
Tong, S. et al. New world bats harbor diverse influenza a viruses. PLoS Pathog. 9, e1003657 (2013).
Air, G. M. Sequence relationships among the hemagglutinin genes of 12 subtypes of influenza A virus. Proc. Natl Acad. Sci. USA 78, 7639–7643 (1981).
Nobusawa, E. et al. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology 182, 475–485 (1991).
Medina, R. A. & Garcia-Sastre, A. Influenza A viruses: new research developments. Nature Rev. Microbiol. 9, 590–603 (2011).
de Graaf, M. & Fouchier, R. A. Role of receptor binding specificity in influenza A virus transmission and pathogenesis. EMBO J. 33, 823–841 (2014).
Wiley, D. C. & Skehel, J. J. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu. Rev. Biochem. 56, 365–394 (1987).
Huang, R. T., Rott, R. & Klenk, H. D. Influenza viruses cause hemolysis and fusion of cells. Virology 110, 243–247 (1981).
Maeda, T. & Ohnishi, S. Activation of influenza virus by acidic media causes hemolysis and fusion of erythrocytes. FEBS Lett. 122, 283–287 (1980).
White, J., Matlin, K. & Helenius, A. Cell fusion by Semliki Forest, influenza, and vesicular stomatitis viruses. J. Cell Biol. 89, 674–679 (1981).
Shaw, M. L., Stone, K. L., Colangelo, C. M., Gulcicek, E. E. & Palese, P. Cellular proteins in influenza virus particles. PLoS Pathog. 4, e1000085 (2008).
Leser, G. P. & Lamb, R. A. Influenza virus assembly and budding in raft-derived microdomains: a quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology 342, 215–227 (2005).
Sauter, N. K. et al. Hemagglutinins from two influenza virus variants bind to sialic acid derivatives with millimolar dissociation constants: a 500-MHz proton nuclear magnetic resonance study. Biochemistry 28, 8388–8396 (1989).
Takemoto, D. K., Skehel, J. J. & Wiley, D. C. A surface plasmon resonance assay for the binding of influenza virus hemagglutinin to its sialic acid receptor. Virology 217, 452–458 (1996).
Gambaryan, A. S. et al. Specification of receptor-binding phenotypes of influenza virus isolates from different hosts using synthetic sialylglycopolymers: non-egg-adapted human H1 and H3 influenza A and influenza B viruses share a common high binding affinity for 6′-sialyl(N-acetyllactosamine). Virology 232, 345–350 (1997).
Eisen, M. B., Sabesan, S., Skehel, J. J. & Wiley, D. C. Binding of the influenza A virus to cell-surface receptors: structures of five hemagglutinin-sialyloligosaccharide complexes determined by X-ray crystallography. Virology 232, 19–31 (1997).
Chu, V. C. & Whittaker, G. R. Influenza virus entry and infection require host cell N-linked glycoprotein. Proc. Natl Acad. Sci. USA 101, 18153–18158 (2004).
de Vries, E. et al. Influenza A virus entry into cells lacking sialylated N-glycans. Proc. Natl Acad. Sci. USA 109, 7457–7462 (2012).
Ablan, S., Rawat, S. S., Blumenthal, R. & Puri, A. Entry of influenza virus into a glycosphingolipid-deficient mouse skin fibroblast cell line. Arch. Virol. 146, 2227–2238 (2001).
Shinya, K. et al. Avian flu: influenza virus receptors in the human airway. Nature 440, 435–436 (2006).
Wilson, I. A., Skehel, J. J. & Wiley, D. C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature 289, 366–373 (1981). This paper provides the first three-dimensional structure of influenza virus HA.
Shi, Y. et al. Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses. Science 342, 243–247 (2013).
Xu, R., McBride, R., Nycholat, C. M., Paulson, J. C. & Wilson, I. A. Structural characterization of the hemagglutinin receptor specificity from the 2009 H1N1 influenza pandemic. J. Virol. 86, 982–990 (2012).
Zhang, W. et al. Molecular basis of the receptor binding specificity switch of the hemagglutinins from both the 1918 and 2009 pandemic influenza A viruses by a D225G substitution. J. Virol. 87, 5949–5958 (2013).
Gamblin, S. J. et al. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science 303, 1838–1842 (2004). References 29, 30 and 31 provide a structural explanation for the receptor-binding shift of H1 subtype viruses.
Liu, J. et al. Structures of receptor complexes formed by hemagglutinins from the Asian Influenza pandemic of 1957. Proc. Natl Acad. Sci. USA 106, 17175–17180 (2009). This paper provides the structural basis of the receptor-binding shift of H2 subtype viruses.
Lin, T. et al. The hemagglutinin structure of an avian H1N1 influenza A virus. Virology 392, 73–81 (2009).
Alonso, M. et al. Resistance and virulence mutations in patients with persistent infection by pandemic 2009 A/H1N1 influenza. J. Clin. Virol. 50, 114–118 (2011).
Reid, A. H., Fanning, T. G., Hultin, J. V. & Taubenberger, J. K. Origin and evolution of the 1918 “Spanish” influenza virus hemagglutinin gene. Proc. Natl Acad. Sci. USA 96, 1651–1656 (1999).
Rogers, G. N. & D'Souza, B. L. Receptor binding properties of human and animal H1 influenza virus isolates. Virology 173, 317–322 (1989).
Cox, N. J. & Subbarao, K. Global epidemiology of influenza: past and present. Annu. Rev. Med. 51, 407–421 (2000).
Pappas, C. et al. Receptor specificity and transmission of H2N2 subtype viruses isolated from the pandemic of 1957. PLoS ONE 5, e11158 (2010).
Matrosovich, M. et al. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J. Virol. 74, 8502–8512 (2000).
Ha, Y., Stevens, D. J., Skehel, J. J. & Wiley, D. C. X-ray structure of the hemagglutinin of a potential H3 avian progenitor of the 1968 Hong Kong pandemic influenza virus. Virology 309, 209–218 (2003).
Lin, Y. P. et al. Evolution of the receptor binding properties of the influenza A(H3N2) hemagglutinin. Proc. Natl Acad. Sci. USA 109, 21474–21479 (2012). References 40 and 41 describe the structural basis of the receptor-binding shift of H3 subtype viruses.
Nobusawa, E., Ishihara, H., Morishita, T., Sato, K. & Nakajima, K. Change in receptor-binding specificity of recent human influenza A viruses (H3N2): a single amino acid change in hemagglutinin altered its recognition of sialyloligosaccharides. Virology 278, 587–596 (2000).
Stevens, J. et al. Receptor specificity of influenza A H3N2 viruses isolated in mammalian cells and embryonated chicken eggs. J. Virol. 84, 8287–8299 (2010).
Owen, R. E. et al. Alterations in receptor binding properties of recent human influenza H3N2 viruses are associated with reduced natural killer cell lysis of infected cells. J. Virol. 81, 11170–11178 (2007).
Asaoka, N. et al. Low growth ability of recent influenza clinical isolates in MDCK cells is due to their low receptor binding affinities. Microbes Infect. 8, 511–519 (2006).
[No authors listed.] Isolation of avian influenza A(H5N1) viruses from humans — Hong Kong, May–December 1997. MMWR Morb. Mortal. Wkly Rep. 46, 1204–1207 (1997).
Stevens, J. et al. Recent avian H5N1 viruses exhibit increased propensity for acquiring human receptor specificity. J. Mol. Biol. 381, 1382–1394 (2008).
Watanabe, Y. et al. Acquisition of human-type receptor binding specificity by new H5N1 influenza virus sublineages during their emergence in birds in Egypt. PLoS Pathog. 7, e1002068 (2011).
Gao, Y. et al. Identification of amino acids in HA and PB2 critical for the transmission of H5N1 avian influenza viruses in a mammalian host. PLoS Pathog. 5, e1000709 (2009).
Herfst, S. et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336, 1534–1541 (2012).
Imai, M. et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486, 420–428 (2012).
Chen, L. M. et al. In vitro evolution of H5N1 avian influenza virus toward human-type receptor specificity. Virology 422, 105–113 (2012).
Zhang, W. et al. An airborne transmissible avian influenza H5 hemagglutinin seen at the atomic level. Science 340, 1463–1467 (2013).
Xiong, X. et al. Receptor binding by a ferret-transmissible H5 avian influenza virus. Nature 497, 392–396 (2013). References 53 and 54 describe the structural basis of the receptor-binding shift in artificial avian influenza H5N1 viruses that have been modified to be transmissible through the air.
Lu, X. et al. Structure and receptor-binding properties of an airborne transmissible avian influenza A virus hemagglutinin H5 (VN1203mut). Protein Cell 4, 502–511 (2013).
Stevens, J. et al. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 312, 404–410 (2006).
Ha, Y., Stevens, D. J., Skehel, J. J. & Wiley, D. C. X-ray structures of H5 avian and H9 swine influenza virus hemagglutinins bound to avian and human receptor analogs. Proc. Natl Acad. Sci. USA 98, 11181–11186 (2001).
Linster, M. et al. Identification, characterization, and natural selection of mutations driving airborne transmission of A/H5N1 virus. Cell 157, 329–339 (2014).
Zhang, Y. et al. H5N1 hybrid viruses bearing 2009/H1N1 virus genes transmit in guinea pigs by respiratory droplet. Science 340, 1459–1463 (2013).
Maher, J. A. & DeStefano, J. The ferret: an animal model to study influenza virus. Lab Anim. 33, 50–53 (2004).
Russell, C. A. et al. The potential for respiratory droplet-transmissible A/H5N1 influenza virus to evolve in a mammalian host. Science 336, 1541–1547 (2012).
Webster, R. G., Geraci, J., Petursson, G. & Skirnisson, K. Conjunctivitis in human beings caused by influenza A virus of seals. N. Engl. J. Med. 304, 911 (1981).
Banks, J., Speidel, E. & Alexander, D. J. Characterisation of an avian influenza A virus isolated from a human — is an intermediate host necessary for the emergence of pandemic influenza viruses? Arch. Virol. 143, 781–787 (1998).
[No authors listed.] Update: influenza activity — United States and worldwide, 2003–2004 season, and composition of the 2004–2005 influenza vaccine. MMWR Morb. Mortal. Wkly Rep. 53, 547–552 (2004).
Fouchier, R. A. et al. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc. Natl Acad. Sci. USA 101, 1356–1361 (2004).
Hirst, M. et al. Novel avian influenza H7N3 strain outbreak, British Columbia. Emerg. Infect. Dis. 10, 2192–2195 (2004).
Koopmans, M. et al. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands. Lancet 363, 587–593 (2004).
Kurtz, J., Manvell, R. J. & Banks, J. Avian influenza virus isolated from a woman with conjunctivitis. Lancet 348, 901–902 (1996).
Nguyen-Van-Tam, J. S. et al. Outbreak of low pathogenicity H7N3 avian influenza in UK, including associated case of human conjunctivitis. Euro Surveill. 11, E060504.2 (2006).
Puzelli, S. et al. Serological analysis of serum samples from humans exposed to avian H7 influenza viruses in Italy between 1999 and 2003. J. Infect. Dis. 192, 1318–1322 (2005).
Tweed, S. A. et al. Human illness from avian influenza H7N3, British Columbia. Emerg. Infect. Dis. 10, 2196–2199 (2004).
Gao, R. et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl. J. Med. 368, 1888–1897 (2013).
Chen, Y. et al. Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: clinical analysis and characterisation of viral genome. Lancet 381, 1916–1925 (2013).
Li, J. et al. Environmental connections of novel avian-origin H7N9 influenza virus infection and virus adaptation to the human. Sci. China Life Sci. 56, 485–492 (2013).
Bao, C. J. et al. Live-animal markets and influenza A (H7N9) virus infection. N. Engl. J. Med. 368, 2337–2339 (2013).
Zhu, H. et al. Infectivity, transmission, and pathology of human-isolated H7N9 influenza virus in ferrets and pigs. Science 341, 183–186 (2013).
Belser, J. A. et al. Pathogenesis and transmission of avian influenza A (H7N9) virus in ferrets and mice. Nature 501, 556–559 (2013).
Watanabe, T. et al. Characterization of H7N9 influenza A viruses isolated from humans. Nature 501, 551–555 (2013).
Richard, M. et al. Limited airborne transmission of H7N9 influenza A virus between ferrets. Nature 501, 560–563 (2013).
Xiong, X. et al. Receptor binding by an H7N9 influenza virus from humans. Nature 499, 496–499 (2013).
Xu, R. et al. Preferential recognition of avian-like receptors in human influenza A H7N9 viruses. Science 342, 1230–1235 (2013).
Yang, H., Carney, P. J., Chang, J. C., Villanueva, J. M. & Stevens, J. Structural analysis of the hemagglutinin from the recent 2013 H7N9 influenza virus. J. Virol. 87, 12433–12446 (2013). References 28, 80, 81 and 82 describe the receptor-binding properties of 2013 human-infecting H7N9 influenza viruses and the structural basis for the host jump.
Cui, L. et al. Dynamic reassortments and genetic heterogeneity of the human-infecting influenza A (H7N9) virus. Nature Commun. 5, 3142 (2014).
Breg, J., Kroon-Batenburg, L. M., Strecker, G., Montreuil, J. & Vliegenthart, J. F. Conformational analysis of the sialyl α(2→3/6)N-acetyllactosamine structural element occurring in glycoproteins, by two-dimensional NOE 1H-NMR spectroscopy in combination with energy calculations by hard-sphere exo-anomeric and molecular mechanics force-field with hydrogen-bonding potential. Eur. J. Biochem. 178, 727–739 (1989).
Poppe, L., Stuike-Prill, R., Meyer, B. & van Halbeek, H. The solution conformation of sialyl-α(2→6)-lactose studied by modern NMR techniques and Monte Carlo simulations. J. Biomol. NMR 2, 109–136 (1992).
Sabesan, S., Bock, K. & Paulson, J. C. Conformational analysis of sialyloligosaccharides. Carbohydr. Res. 218, 27–54 (1991).
Stehle, T. & Harrison, S. C. Crystal structures of murine polyomavirus in complex with straight-chain and branched-chain sialyloligosaccharide receptor fragments. Structure 4, 183–194 (1996).
Wright, C. S. & Jaeger, J. Crystallographic refinement and structure analysis of the complex of wheat germ agglutinin with a bivalent sialoglycopeptide from glycophorin A. J. Mol. Biol. 232, 620–638 (1993).
Merritt, E. A. et al. Crystal structure of cholera toxin B-pentamer bound to receptor GM1 pentasaccharide. Protein Sci. 3, 166–175 (1994).
Lam, T. T. et al. The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature 502, 241–244 (2013).
Couceiro, J. N., Paulson, J. C. & Baum, L. G. Influenza virus strains selectively recognize sialyloligosaccharides on human respiratory epithelium; the role of the host cell in selection of hemagglutinin receptor specificity. Virus Res. 29, 155–165 (1993).
Webster, R. G., Bean, W. J., Gorman, O. T., Chambers, T. M. & Kawaoka, Y. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56, 152–179 (1992).
Wise, H. M. et al. A complicated message: identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. J. Virol. 83, 8021–8031 (2009).
Jagger, B. W. et al. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 337, 199–204 (2012).
Acknowledgements
Work in the authors' laboratory was supported by the China Ministry of Science and Technology National 973 Project (grant no. 2011CB504703), the National Natural Science Foundation of China (NSFC; grant no. 81290342 and 31402196) and intramural special grant for influenza virus research from the Chinese Academy of Sciences (KJZD-EW-L09). G.F.G. is a leading principal investigator of the NSFC Innovative Research Group (grant no. 81321063). Y.S. is supported by the Excellent Young Scientist Program of the Chinese Academy of Sciences. The authors are grateful to B. Tefsen and J. Haywood for their help in preparing the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- Herd immunity
-
(Also known as herd effect or community immunity). A form of immunity that occurs when the vaccination of a substantial portion of a population (or herd) provides a measure of protection for individuals who have not developed immunity.
- Macropinocytic endocytosis
-
A process by which cells internalize molecules using a non-selective uptake pattern.
- Clathrin-mediated endocytosis
-
A process by which cells internalize molecules in a receptor-mediated uptake pattern; it is also called receptor-mediated endocytosis.
- Trans conformation
-
The conformation of a glycosidic linkage of receptor analogues, in which the two carbon atoms at either end of the glycosidic bond are oriented in opposing directions.
- Cis conformation
-
The conformation of a glycosidic linkage of a receptor analogue, in which the two carbon atoms at either end of the glycosidic bond are oriented in the same direction.
- Surface plasmon resonance
-
A method used to measure the interaction of macromolecules at a surface via changes in the refractive index.
- Microarray
-
A two-dimensional array on a solid substrate (usually a glass or plastic slide) that numerous biomaterials are attached to in a regular pattern for biochemical or genetic analysis.
- Reassortant
-
A term that usually refers to a virus that contains two or more segments of nucleic acid (segmented genome) that are derived from different viruses.
Rights and permissions
About this article
Cite this article
Shi, Y., Wu, Y., Zhang, W. et al. Enabling the 'host jump': structural determinants of receptor-binding specificity in influenza A viruses. Nat Rev Microbiol 12, 822–831 (2014). https://doi.org/10.1038/nrmicro3362
Published:
Issue date:
DOI: https://doi.org/10.1038/nrmicro3362
This article is cited by
-
Surveillance and characterization of avian-origin H3N2 canine influenza viruses in 2021 in China
One Health Advances (2024)
-
Genetic characterization and pathogenicity of a Eurasian avian-like H1N1 swine influenza reassortant virus
Virology Journal (2022)
-
An epitope-optimized human H3N2 influenza vaccine induces broadly protective immunity in mice and ferrets
npj Vaccines (2022)
-
Structural basis for a human broadly neutralizing influenza A hemagglutinin stem-specific antibody including H17/18 subtypes
Nature Communications (2022)
-
Blood group-gut microbiome—health axis gains further support from landmark multi-omics study in swines
Science China Life Sciences (2022)