Abstract
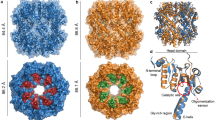
SecB is a bacterial molecular chaperone involved in mediating translocation of newly synthesized polypeptides across the cytoplasmic membrane of bacteria. The crystal structure of SecB from Haemophilus influenzae shows that the molecule is a tetramer organized as a dimer of dimers. Two long channels run along the side of the molecule. These are bounded by flexible loops and lined with conserved hydrophobic amino acids, which define a suitable environment for binding non-native polypeptides. The structure also reveals an acidic region on the top surface of the molecule, several residues of which have been implicated in binding to SecA, its downstream target.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
von Heijne, G. The signal peptide. J. Membr. Biol. 115, 195–201 (1990).
Trun, N.J., Stader, J., Lupas, A., Kumamoto, C. & Silhavy, T.J. Two cellular components, PrlA and SecB, that recognize different sequence determinants are required for efficient protein export. J. Bacteriol. 170, 5928– 5930 (1988).
Lecker, S. et al. Three pure chaperone proteins of Escherichia coli, SecB, trigger factor and GroEL, form soluble complexes with precursor proteins in vitro. EMBO J. 8, 2703– 2709 (1989).
Kumamoto, C A. Escherichia coli SecB protein associates with exported protein precursors in vivo. Proc. Natl. Acad. Sci. USA 86, 5320–5324 (1989).
Randall, L.L. et al. Binding of SecB to ribosome-bound polypeptides has the same characteristics as binding to full-length, denatured proteins. Proc. Natl. Acad. Sci. USA 94, 802– 807 (1997).
Hartl, F.U., Lecker, S., Schiebel, E., Hendrick, J.P. & Wickner, W. The binding cascade of SecB to SecA to SecY/E mediates preprotein targeting to the E. coli plasma membrane. Cell 63, 269– 279 (1990).
Fekkes, P. et al. Preprotein transfer to the Escherichia coli translocase requires the co-operative binding of SecB and the signal sequence to SecA . Mol. Microbiol. 29, 1179– 1190 (1998).
Hardy, S.J. & Randall, L.L. Recognition of ligands by SecB, a molecular chaperone involved in bacterial protein export. Philos. Trans. R. Soc. Lond. B Biol. Sci. 339, 343– 352; discussion 352–344 ( 1993).
Kumamoto, C.A. & Beckwith, J. Evidence for specificity at an early step in protein export in Escherichia coli. J. Bacteriol. 163, 267–274 (1985).
Weiss, J.B., Ray, P.H. & Bassford, P.J., Jr. . Purified secB protein of Escherichia coli retards folding and promotes membrane translocation of the maltose-binding protein in vitro. Proc. Natl. Acad. Sci. USA 85, 8978–8982 (1988).
Chen, L. & Sigler, P.B. The crystal structure of a GroEL/peptide complex: plasticity as a basis for substrate diversity . Cell 99, 757–768 (1999).
Zhu, X. et al. Structural analysis of substrate binding by the molecular chaperone DnaK. Science 272, 1606– 1614 (1996).
Randall, L.L. Peptide binding by chaperone SecB: implications for recognition of nonnative structure. Science 257, 241– 245 (1992).
Kumamoto, C.A. & Francetic, O. Highly selective binding of nascent polypeptides by an Escherichia coli chaperone protein in vivo. J. Bacteriol. 175, 2184– 2188 (1993).
Volkert, T.L., Baleja, J.D. & Kumamoto, C.A. A highly mobile C-terminal tail of the Escherichia coli protein export chaperone SecB. Biochem. Biophys. Res. Commun. 264, 949–954 (1999).
Kimsey, H.H., Dagarag, M.D. & Kumamoto, C.A. Diverse effects of mutation on the activity of the Escherichia coli export chaperone SecB. J. Biol. Chem. 270, 22831–22835 (1995).
Muren, E.M., Suciu, D., Topping, T.B., Kumamoto, C.A. & Randall, L.L. Mutational alterations in the homotetrameric chaperone SecB that implicate the structure as dimer of dimers. J. Biol. Chem. 274, 19397– 19402 (1999).
Panse, V.G., Swaminathan, C.P., Surolia, A. & Varadarajan, R. Thermodynamics of substrate binding to the chaperone SecB. Biochemistry 39, 2420–2427 ( 2000).
Randall, L.L., Topping, T.B. & Hardy, S.J. No specific recognition of leader peptide by SecB, a chaperone involved in protein export. Science 248, 860–863 (1990).
Randall, L.L., Topping, T.B., Suciu, D. & Hardy, S.J. Calorimetric analyses of the interaction between SecB and its ligands. Protein Sci. 7, 1195–1200 (1998).
Eck, M.J., Dhe-Paganon, S., Trub, T., Nolte, R.T., Shoelson, S. E. Structure of the IRS-1 PTB domain bound to the juxtamembrane region of the insulin receptor. Cell 85, 695–705 ( 1996).
Kavanaugh, W.M., Turck, C.W., Williams, L.T. PTB domain binding to signaling proteins through a sequence motif containing phosphotyrosine. Science 268, 1177–1179 (1995).
Knoblauch, N.T., et al. Substrate specificity of the SecB chaperone. J. Biol. Chem. 274, 34219–34225 (1999).
Smith, V.F., Schwartz, B.L., Randall, L.L. & Smith, R.D. Electrospray mass spectrometric investigation of the chaperone SecB. Protein Sci. 5, 488–494 (1996).
Wickner, W. & Leonard, M.R. Escherichia coli preprotein translocase. J. Biol. Chem. 271, 29514–29516 (1996).
Woodbury, R.L. et al. Complexes between protein export chaperone SecB and SecA. Evidence for separate sites on SecA providing binding energy and regulatory interactions. J. Biol. Chem. 275, 24191– 24198 (2000).
Fekkes, P., van der Does, C. & Driessen, A.J. The molecular chaperone SecB is released from the carboxy-terminus of SecA during initiation of precursor protein translocation . EMBO J. 16, 6105–6113 (1997).
Altman, E., Kumamoto, C.A. & Emr, S.D. Heat-shock proteins can substitute for SecB function during protein export in Escherichia coli. EMBO J. 10, 239–245 (1991).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 ( 1997).
Hendrickson, W.A. & Ogata, C.M. Phase determination from multiwavelength anomalous diffraction measurements. Methods Enzymol. 276, 494–523 ( 1997).
Brünger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Thompson, J.D., Higgins, D.G., T.J. CLUSTAL W: improving the sensitivity of progressive multiple squence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673– 4680 (1994).
Kraulis, P.J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946 –950 (1991).
The persistance of vision raytracer version 3.1g, POV-team, Williamstown, Victoria, Australia. http://www.povray.org
Nicholls, A., Sharp, K.A. & Honig, B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11, 281–296 (1991).
Acknowledgements
We thank J. Stuckey for maintaining the X-ray facility at the University of Michigan Medical School and for assistance on synchrotron data collection; M. Marletta and D. Peisach for access to their SGI graphic workstation during structure fitting and refinement; K. Brister for access and help at APS BioCARs beamlines; J. Dixon, M. Marletta, K. Orth and R. Taussig for critically reading the manuscript. This work was supported by an NIH grant to Z.X. and the University of Michigan Biological Scholar Program.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xu, Z., Knafels, J. & Yoshino, K. Crystal structure of the bacterial protein export chaperone SecB. Nat Struct Mol Biol 7, 1172–1177 (2000). https://doi.org/10.1038/82040
Received:
Accepted:
Issue date:
DOI: https://doi.org/10.1038/82040
This article is cited by
-
Molecular chaperones and their denaturing effect on client proteins
Journal of Biomolecular NMR (2021)
-
Structural insights into chaperone addiction of toxin-antitoxin systems
Nature Communications (2019)
-
The Principles of Protein Targeting and Transport Across Cell Membranes
The Protein Journal (2019)
-
Protein Transport Across the Bacterial Plasma Membrane by the Sec Pathway
The Protein Journal (2019)
-
Protein export through the bacterial Sec pathway
Nature Reviews Microbiology (2017)