Abstract
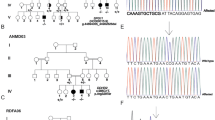
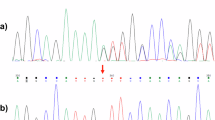
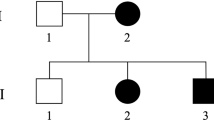
Hereditary spastic paraplegias are a group of genetically heterogeneous neurological disorders characterized by progressive weakness and spasticity of lower limbs. We ascertained five families with eight individuals with hereditary spastic paraplegia. Pathogenic variants were identified by exome sequencing of index cases. The cohort consists of three families with spastic paraplegia type 47 (AP4B1) with a common mutation in two families, a family with spastic paraplegia type 50 (AP4M1), and two male siblings with X-linked spastic paraplegia 2 (PLP1). This work illustrates locus and allelic heterogeneity in five families with hereditary spastic paraplegia.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Klebe S, Stevanin G, Depienne C. Clinical and genetic heterogeneity in hereditary spastic paraplegias: from SPG1 to SPG72 and still counting. Rev Neurol (Paris). 2015;171:505–30.
Ruano L, Melo C, Silva MC, Coutinho P. The global epidemiology of hereditary ataxia and spastic paraplegia: a systematic review of prevalence studies. Neuroepidemiology. 2014;42:174–83.
de Souza PV, de Rezende Pinto WB, de Rezende Batistella GN, Bortholin T, Oliveira AS. Hereditary spastic paraplegia: clinical and genetic hallmarks. Cerebellum. 2017;16:525–51.
Denton KR, Xu C, Shah H, Li XJ. Modeling axonal defects in hereditary spastic paraplegia with human pluripotent stem cells. Front Biol. 2016;11:339–54.
Girisha KM, Shukla A, Trujillano D, Bhavani GS, Hebbar M, Kadavigere R, et al. A homozygous nonsense variant in IFT52 is associated with a human skeletal ciliopathy. Clin Genet. 2016;90:536–9.
Guo Y, Ding X, Shen Y, Lyon GJ, Wang K. SeqMule: automated pipeline for analysis of human exome/genome sequencing data. Sci Rep. 2015;5:14283.
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, et al. The variant call format and VCFtools. Bioinforma (Oxf, Engl). 2011;27:2156–8.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med: Off J Am Coll Med Genet. 2015;17:405–24.
Hand CK, Bernard G, Dube MP, Shevell MI, Rouleau GA. A novel PLP1 mutation further expands the clinical heterogeneity at the locus. Can J Neurol Sci Le J Can Des Sci Neurol. 2012;39:220–4.
Abou Jamra R, Philippe O, Raas-Rothschild A, Eck SH, Graf E, Buchert R, et al. Adaptor protein complex 4 deficiency causes severe autosomal-recessive intellectual disability, progressive spastic paraplegia, shy character, and short stature. Am J Hum Genet. 2011;88:788–95.
Bauer P, Leshinsky-Silver E, Blumkin L, Schlipf N, Schroder C, Schicks J, et al. Mutation in the AP4B1 gene cause hereditary spastic paraplegia type 47 (SPG47). Neurogenetics. 2012;13:73–76.
Abdollahpour H, Alawi M, Kortum F, Beckstette M, Seemanova E, Komarek V, et al. An AP4B1 frameshift mutation in siblings with intellectual disability and spastic tetraplegia further delineates the AP-4 deficiency syndrome. Eur J Human Genet: Ejhg. 2015;23:256–9.
Tuysuz B, Bilguvar K, Kocer N, Yalcinkaya C, Caglayan O, Gul E, et al. Autosomal recessive spastic tetraplegia caused by AP4M1 and AP4B1 gene mutation: expansion of the facial and neuroimaging features. Am J Med Genet A. 2014;164a:1677–85.
Ebrahimi-Fakhari D, Cheng C, Dies K, Diplock A, Pier DB, Ryan CS, et al. Clinical and genetic characterization of AP4B1-associated SPG47. Am J Med Genet A. 2018;176:311–8.
Bettencourt C, Salpietro V, Efthymiou S, Chelban V, Hughes D, Pittman AM, et al. Genotype-phenotype correlations and expansion of the molecular spectrum of AP4M1-related hereditary spastic paraplegia. Orphanet J Rare Dis. 2017;12:172.
Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–64.
Verkerk AJ, Schot R, Dumee B, Schellekens K, Swagemakers S, Bertoli-Avella AM, et al. Mutation in the AP4M1 gene provides a model for neuroaxonal injury in cerebral palsy. Am J Hum Genet. 2009;85:40–52.
Langouet M, Siquier-Pernet K, Sanquer S, Bole-Feysot C, Nitschke P, Boddaert N, et al. Contiguous mutation syndrome in the era of high-throughput sequencing. Mol Genet & Genom Med. 2015;3:215–20.
Jameel M, Klar J, Tariq M, Moawia A, Altaf Malik N, Seema Waseem S, et al. A novel AP4M1 mutation in autosomal recessive cerebral palsy syndrome and clinical expansion of AP-4 deficiency. BMC Med Genet. 2014;15:133.
Najmabadi H, Hu H, Garshasbi M, Zemojtel T, Abedini SS, Chen W, et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature. 2011;478:57–63.
Alazami AM, Patel N, Shamseldin HE, Anazi S, Al-Dosari MS, Alzahrani F, et al. Accelerating novel candidate gene discovery in neurogenetic disorders via whole-exome sequencing of prescreened multiplex consanguineous families. Cell Rep. 2015;10:148–61.
Acknowledgements
We thank the families who cooperated with evaluation of their children and consented for participation in this study.
Funding
This work was supported by Department of Health Research funded the project titled “Clinical and molecular characterization of leukodystrophies in Indian children” (V.25011/379/2015-GIA/HR) and National Institutes of Health funded project, ‘Genetic Diagnosis of Heritable Neurodevelopmental Disorders in India: Investigating the Use of Whole Exome Sequencing and Genetic Counseling to Address the High Burden of Neurodevelopmental Disorders’(1R21NS094047-01).
Author information
Authors and Affiliations
Contributions
KMG and AS evaluated the patients, conceived the idea of the manuscript and planned the experiments. MH performed experiments and drafted the manuscript. SN evaluated the patients and contributed to the clinical reports. SB and KMG supervised the entire work. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Hebbar, M., Shukla, A., Nampoothiri, S. et al. Locus and allelic heterogeneity in five families with hereditary spastic paraplegia. J Hum Genet 64, 17–21 (2019). https://doi.org/10.1038/s10038-018-0523-y
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s10038-018-0523-y
This article is cited by
-
A novel loss of function mutation in adaptor protein complex 4, subunit mu-1 causing autosomal recessive spastic paraplegia 50
Neurological Sciences (2021)
-
Novel variants in AP4B1 cause spastic tetraplegia, moderate psychomotor development delay and febrile seizures in a Chinese patient: a case report
BMC Medical Genetics (2020)