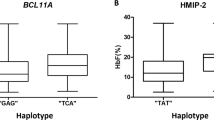
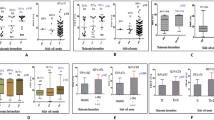
Abstract
β-S globin haplotype (βS haplotype) characterization in sickle cell anemia (SCA) patients is important because it assists individualized treatment. However, the patient with atypical haplotypes do not present detailed studies such as clinical and laboratory data. To understand the phenotypic expression of atypical haplotype patients in relation to typical haplotype ones, it may be necessary to assess the main clinical and laboratorial parameters and investigate transcription factors, as possible genetic modulators that can contribute to the improvement of the SCA patients’ clinical condition. The study group was composed of 600 SCA Brazilian patients of both genders ranging in age from 1 to 68 years. The atypical haplotypes were the third most frequent (5.7%) with 11 patterns numerically ranked according to occurrence. We verified that patients with atypical 1 haplotype in combination with Bantu haplotype presented milder clinical outcomes in relation to Bantu/Bantu and Benin/Benin patients, according to improved values of hemoglobin and hematocrit. In clinical severity, we did not observe significant statistical differences between typical and atypical haplotype patients, and this result can be explained with reference to the action of transcription factors in β-globin cluster. Thus, we presented the atypical haplotype relationship with SCA pathophysiology, reinforcing the hypothesis that individual genetic factors may be responsible for phenotypic diversity of the disease.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Rees DC, Williams TN, Gladwin MT. Sickle-cell disease. Lancet. 2010;376:2018–31.
Ingram VM. Gene mutations in human haemoglobin: the chemical difference between normal and sickle cell haemoglobin. Nature. 1957;180:326–8.
Kan YW, Dozy M. Polymorphism of DNA sequence adjacent to human beta-globin structural gene: relationship to sickle mutation. Proc Natl Acad Sci USA. 1978;75:5631–5.
Mears JG, et al. Its origin and diffusion from West Africa. J Clin Invest. 1981;68:606–10.
Pagnier J, et al. Evidence for the multicentric origin of the sickle cell hemoglobin gene in Africa. Proc Natl Acad Sci USA. 1984;81:1771–3.
Hockham C, Piel FB, Gupta S, Penman BS. Understanding the contrasting spatial haplotype patterns of malaria-protective beta-globin polymorphisms. Infect Genet Evol. 2015;36:174–83.
Kulozik aE, et al. Geographical survey of beta S-globin gene haplotypes: evidence for an independent Asian origin of the sickle-cell mutation. Am J Hum Genet. 1986;39:239–44.
Lapouméroulie C, et al. A novel sickle cell mutation of yet another origin in Africa: the Cameroon type. Hum Genet. 1992;89:333–7.
Srinivas R, et al. Atypical haplotypes linked to the beta-S gene in Africa are likely to be the product of recombination. Am J Hematol. 1988;29:60–62.
Zago Ma, et al. Atypical beta(s) haplotypes are generated by diverse genetic mechanisms. Am J Hematol. 2000;63:79–84.
Nagel RL. The origin of the hemoglobin S gene: clinical, genetic and anthropological consequences. Einst Q J Biol Med. 1984;2:53–62.
Gabriel A, Przybylski J. Sickle cell anaemia: a look at global haplotype distribution. Nat Educ. 2010;3:2.
Sambrook J, Fritsch EF, Maniatis, T MolecularCloning: a laboratory manual. Cold Spring Harb Lab Press 1989. http://www.molecularcloning.com/.
Saiki RK, et al. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science (80-). 1985;230:1350–4.
Sutton M, Bouhassira EE, Nagel RL. Polymerase chain reaction amplification applied to the determination of beta-like globin gene cluster haplotypes. Am J Hematol. 1989;32:66–69.
Nouraie M, et al. The relationship between the severity of hemolysis, clinical manifestations and risk of death in 415 patients with sickle cell anemia in the US and Europe. Haematologica. 2013;98:464–72.
Sebastiani P, et al. A network model to predict the risk of death in sickle cell disease. Blood. 2007;110:2727–35.
Belini Junior E, et al. Severity of Brazilian sickle cell disease patients: severity scores and feasibility of the Bayesian network model use. Blood Cells Mol Dis. 2015;54:321–7.
Mathelier A, et al. JASPAR 2014: An extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42:142–7.
Sandelin A, Alkema W, Engström P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–4.
Hanchard N, et al. Classical sickle beta-globin haplotypes exhibit a high degree of long-range haplotype similarity in African and Afro-Caribbean populations. BMC Genet. 2007;8:52.
Chakravarti a, et al. Nonuniform recombination within the human beta-globin gene cluster. Am J Hum Genet. 1984;36:1239–58.
Powars DR. Beta s-gene-cluster haplotypes in sickle cell anemia. Clinical and hematologic features. Hematol Oncol Clin N Am. 1991;5:475–93.
Powars D, Hiti A. Sickle cell anemia. Beta s gene cluster haplotypes as genetic markers for severe disease expression. Am J Dis Child. 1993;147:1197–202.
Silva-Rocha LB, Dias-Elias DB, Barbosa MC, Bandeira ICJ, Gonçalves RP. DNA damage in leukocytes of sickle cell anemia patients is associated with hydroxyurea therapy and with HBB*S haplotype. Mutat Res Toxicol Environ Mutagen. 2012;749:48–52.
Steinberg MH, et al. Fetal hemoglobin in sickle cell anemia: determinants of response to hydroxyurea. Multicenter Study of Hydroxyurea. Blood. 1997;89:1078–88.
Nagel RL, et al. The hematologic characteristics of sickle cell anemia bearing the Bantu haplotype: the relationship between G-y and Hb F levels. Blood. 1987;69:1026–30.
Zhou W, et al. The role of p22 NF-E4 in human globin gene switching. J Biol Chem. 2004;279:26227–32.
Carrocini GC, de S, Venancio LPR, Bonini-Domingos CR. Screening of transcription factors involved in fetal hemoglobin regulation using phylogenetic footprinting. Evol Bioinforma. 2015;11:239–44.
Soza-ried C, Hess I, Netuschil N, Schorpp M, Boehm T. Essential role of c-myb in definitive hematopoiesis is evolutionarily conserved. Proc Natl Acad Sci USA. 2010;107:17340–08.
Bauer D, Kamram S, Orkim S. Reawakening fetal hemoglobin: prospects for new therapies for the β-globin disorders. Blood. 2012;120:2945–53.
Ikonomi P, et al. Levels of GATA-1/GATA-2 transcription factors modulate expression of embryonic and fetal hemoglobins. Gene. 2000;261:277–87.
Xu J, et al. Transcriptional silencing of beta-globin by BCL11A involves long-range interactions and cooperation with SOX6. Genes Dev. 2010;24:783–9.
Zhu X et al. NF-Y recruits both transcription activator and repressor to modulate tissue- and developmental stage-specific expression of human y-globin gene. PLoS ONE 2012;7: 1–15.
Acknowledgements
We would like to thank Izabella Luisa Tambones (Department of Biology, São Paulo State University – UNESP) for her technical assistance in transcription factors analyses and Joseph People (Department of Physiology, Development and Neuroscience, University of Cambridge) for English proofreading. The Pro-Institute of Hematology Foundation (Fundarj) and the State of Sao Paulo Research Foundation (FAPESP process number 2011/14168-5) for the financial support. Silva is a partnership member of Redoxoma – CEPID/FAPESP (2013/07937-8).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Okumura, J.V., Silva, D.G.H., Torres, L.S. et al. Atypical β-S haplotypes: classification and genetic modulation in patients with sickle cell anemia. J Hum Genet 64, 239–248 (2019). https://doi.org/10.1038/s10038-018-0554-4
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s10038-018-0554-4