Abstract
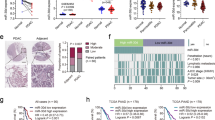
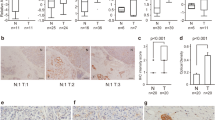
Our ongoing analyses identifying dysregulated microRNAs (miRNAs) and their controlled target RNAs have shed light on novel oncogenic pathways in pancreatic ductal adenocarcinoma (PDAC). The PDAC miRNA signature obtained by RNA sequencing showed that both strands of pre-miR-130b (miR-130b-5p, the passenger strand and miR-130b-3p, the guide strand) were significantly downregulated in cancer tissues. Our functional assays revealed that miR-130b-5p significantly blocked the malignant abilities of PDAC cell lines (PANC-1 and SW1990), e.g., cancer cell proliferation, migration, and invasion. A total of 103 genes were identified as possible oncogenic targets by miR-130b-5p regulation in PDAC cells based on genome-wide gene expression analysis and in silico database search. Among the possible targets, high expression of 9 genes (EPS8, ZWINT, SMC4, LDHA, GJB2, ZCCHC24, TOP2A, ANLN, and ADCY3) predicted a significantly poorer prognosis of PDAC patients (5-year overall survival, p < 0.001). Furthermore, we focused on EPS8 because its expression had the greatest impact on patient prognosis (overall survival, p < 0.0001). Overexpression of EPS8 was detected in PDAC clinical specimens. Knockdown assays with siEPS8 showed that its overexpression enhanced cancer cell proliferation, migration, and invasion. Analysis of downstream RNA networks regulated by EPS8 indicated that MET, HMGA2, FERMT1, RARRES3, PTK2, MAD2L1, and FLI1 were closely involved in PDAC pathogenesis. Genes regulated by antitumor miR-130b-5p were closely involved in PDAC molecular pathogenesis. Our approach, discovery of antitumor miRNAs and their target RNAs, will contribute to exploring the causes of this malignant disease.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388:73–85.
Das S, Batra SK. Pancreatic cancer metastasis: are we being pre-EMTed? Curr Pharm Des. 2015;21:1249–55.
Polireddy K, Chen Q. Cancer of the pancreas: molecular pathways and current advancement in treatment. J Cancer. 2016;7:1497–514.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell . 2004;116:281–97.
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell . 2009;136:215–33.
Liu WW, Meng J, Cui J, Luan YS. Characterization and function of microRNA(*)s in plants. Front Plant Sci. 2017;8:2200.
Goto Y, Kurozumi A, Enokida H, Ichikawa T, Seki N. Functional significance of aberrantly expressed microRNAs in prostate cancer. Int J Urol. 2015;22:242–52.
Koshizuka K, Hanazawa T, Arai T, Okato A, Kikkawa N, Seki N. Involvement of aberrantly expressed microRNAs in the pathogenesis of head and neck squamous cell carcinoma. Cancer Metastas- Rev. 2017;36:525–45.
Yonemori K, Kurahara H, Maemura K, Natsugoe S. MicroRNA in pancreatic cancer. J Hum Genet. 2017;62:33–40.
Itesako T, Seki N, Yoshino H, Chiyomaru T, Yamasaki T, Hidaka H, et al. The microRNA expression signature of bladder cancer by deep sequencing: the functional significance of the miR-195/497 cluster. PLoS ONE. 2014;9:e84311.
Goto Y, Kurozumi A, Arai T, Nohata N, Kojima S, Okato A, et al. Impact of novel miR-145-3p regulatory networks on survival in patients with castration-resistant prostate cancer. Br J Cancer. 2017;117:409–20.
Koshizuka K, Nohata N, Hanazawa T, Kikkawa N, Arai T, Okato A, et al. Deep sequencing-based microRNA expression signatures in head and neck squamous cell carcinoma: dual strands of pre-miR-150 as antitumor miRNAs. Oncotarget . 2017;8:30288–304.
Mizuno K, Mataki H, Arai T, Okato A, Kamikawaji K, Kumamoto T, et al. The microRNA expression signature of small cell lung cancer: tumor suppressors of miR-27a-5p and miR-34b-3p and their targeted oncogenes. J Hum Genet. 2017;62:671–78.
Yonemori K, Seki N, Idichi T, Kurahara H, Osako Y, Koshizuka K, et al. The microRNA expression signature of pancreatic ductal adenocarcinoma by RNA sequencing: anti-tumour functions of the microRNA-216 cluster. Oncotarget . 2017;8:70097–115.
Toda H, Kurozumi S, Kijima Y, Idichi T, Shinden Y, Yamada Y, et al. Molecular pathogenesis of triple-negative breast cancer based on microRNA expression signatures: antitumor miR-204-5p targets AP1S3. J Hum Genet. 2018;63:1197–210.
Yonemori K, Seki N, Kurahara H, Osako Y, Idichi T, Arai T, et al. ZFP36L2 promotes cancer cell aggressiveness and is regulated by antitumor microRNA-375 in pancreatic ductal adenocarcinoma. Cancer Sci. 2017;108:124–35.
Idichi T, Seki N, Kurahara H, Yonemori K, Osako Y, Arai T, et al. Regulation of actin-binding protein ANLN by antitumor miR-217 inhibits cancer cell aggressiveness in pancreatic ductal adenocarcinoma. Oncotarget . 2017;8:53180–93.
Idichi T, Seki N, Kurahara H, Fukuhisa H, Toda H, Shimonosono M, et al. Molecular pathogenesis of pancreatic ductal adenocarcinoma: Impact of passenger strand of pre-miR-148a on gene regulation. Cancer Sci. 2018;109:2013–26.
Idichi T, Seki N, Kurahara H, Fukuhisa H, Toda H, Shimonosono M, et al. Involvement of anti-tumor miR-124-3p and its targets in the pathogenesis of pancreatic ductal adenocarcinoma: direct regulation of ITGA3 and ITGB1 by miR-124-3p. Oncotarget . 2018;9:28849–65.
Yamada Y, Arai T, Sugawara S, Okato A, Kato M, Kojima S, et al. Impact of novel oncogenic pathways regulated by antitumor miR-451a in renal cell carcinoma. Cancer Sci. 2018;109:1239–53.
Yamada Y, Sugawara S, Arai T, Kojima S, Kato M, Okato A, et al. Molecular pathogenesis of renal cell carcinoma: Impact of the anti-tumor miR-29 family on gene regulation. Int J Urol. 2018;25:953–65.
Osako Y, Seki N, Koshizuka K, Okato A, Idichi T, Arai T, et al. Regulation of SPOCK1 by dual strands of pre-miR-150 inhibit cancer cell migration and invasion in esophageal squamous cell carcinoma. J Hum Genet. 2017;62:935–44.
Sugawara S, Yamada Y, Arai T, Okato A, Idichi T, Kato M, et al. Dual strands of the miR-223 duplex (miR-223-5p and miR-223-3p) inhibit cancer cell aggressiveness: targeted genes are involved in bladder cancer pathogenesis. J Hum Genet. 2018;63:657–68.
Yamada Y, Arai T, Kojima S, Sugawara S, Kato M, Okato A, et al. Regulation of antitumor miR-144-5p targets oncogenes: Direct regulation of syndecan-3 and its clinical significance. Cancer Sci. 2018;109:2919–36.
Arai T, Kojima S, Yamada Y, Sugawara S, Kato M, Yamazaki K, et al. Pirin: a potential novel therapeutic target for castration-resistant prostate cancer regulated by miR-455-5p. Mol Oncol. 2019;13:322–37.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1.
Anaya J. OncoLnc: linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Comput Sci. 2016;2:e67.
Mah SM, Buske C, Humphries RK, Kuchenbauer F. miRNA*: a passenger stranded in RNA-induced silencing complex? Crit Rev Eukaryot Gene Expr. 2010;20:141–8.
Yang J, Zeng Y. Identification of miRNA-mRNA crosstalk in pancreatic cancer by integrating transcriptome analysis. Eur Rev Med Pharmacol Sci. 2015;19:825–34.
Yang C, Cai J, Wang Q, Tang H, Cao J, Wu L, et al. Epigenetic silencing of miR-130b in ovarian cancer promotes the development of multidrug resistance by targeting colony-stimulating factor 1. Gynecol Oncol. 2012;124:325–34.
Egawa H, Jingushi K, Hirono T, Ueda Y, Kitae K, Nakata W, et al. The miR-130 family promotes cell migration and invasion in bladder cancer through FAK and Akt phosphorylation by regulating PTEN. Sci Rep. 2016;6:20574.
Ramalho-Carvalho J, Graca I, Gomez A, Oliveira J, Henrique R, Esteller M, et al. Downregulation of miR-130b~301b cluster is mediated by aberrant promoter methylation and impairs cellular senescence in prostate cancer. J Hematol Oncol. 2017;10:43.
Yu T, Cao R, Li S, Fu M, Ren L, Chen W, et al. MiR-130b plays an oncogenic role by repressing PTEN expression in esophageal squamous cell carcinoma cells. BMC Cancer. 2015;15:29.
Zhang Q, Zhang B, Sun L, Yan Q, Zhang Y, Zhang Z, et al. MicroRNA-130b targets PTEN to induce resistance to cisplatin in lung cancer cells by activating Wnt/beta-catenin pathway. Cell Biochem Funct. 2018;36:194–202.
Dong P, Karaayvaz M, Jia N, Kaneuchi M, Hamada J, Watari H, et al. Mutant p53 gain-of-function induces epithelial-mesenchymal transition through modulation of the miR-130b-ZEB1 axis. Oncogene . 2013;32:3286–95.
Dettmer MS, Perren A, Moch H, Komminoth P, Nikiforov YE, Nikiforova MN. MicroRNA profile of poorly differentiated thyroid carcinomas: new diagnostic and prognostic insights. J Mol Endocrinol. 2014;52:181–9.
Zhao G, Zhang JG, Shi Y, Qin Q, Liu Y, Wang B, et al. MiR-130b is a prognostic marker and inhibits cell proliferation and invasion in pancreatic cancer through targeting STAT3. PLoS ONE. 2013;8:e73803.
Fazioli F, Minichiello L, Matoska V, Castagnino P, Miki T, Wong WT, et al. Eps8, a substrate for the epidermal growth factor receptor kinase, enhances EGF-dependent mitogenic signals. EMBO J. 1993;12:3799–808.
Logue JS, Cartagena-Rivera AX, Baird MA, Davidson MW, Chadwick RS, Waterman CM. Erk regulation of actin capping and bundling by Eps8 promotes cortex tension and leader bleb-based migration. eLife . 2015;4:e08314.
Chen H, Wu X, Pan ZK, Huang S. Integrity of SOS1/EPS8/ABI1 tri-complex determines ovarian cancer metastasis. Cancer Res. 2010;70:9979–90.
Maa MC, Lee JC, Chen YJ, Chen YJ, Lee YC, Wang ST, et al. Eps8 facilitates cellular growth and motility of colon cancer cells by increasing the expression and activity of focal adhesion kinase. J Biol Chem. 2007;282:19399–409.
Chen C, Liang Z, Huang W, Li X, Zhou F, Hu X, et al. Eps8 regulates cellular proliferation and migration of breast cancer. Int J Oncol. 2015;46:205–14.
He YZ, Liang Z, Wu MR, Wen Q, Deng L, Song CY, et al. Overexpression of EPS8 is associated with poor prognosis in patients with acute lymphoblastic leukemia. Leuk Res. 2015;39:575–81.
Li Q, Bao W, Fan Q, Shi WJ, Li ZN, Xu Y, et al. Epidermal growth factor receptor kinase substrate 8 promotes the metastasis of cervical cancer via the epithelial-mesenchymal transition. Mol Med Rep. 2016;14:3220–8.
Acknowledgements
This study was supported by Japan Society for the Promotion of Science 17H04285, 18K08626, 18K08687, 18K16322, 18K09338, 16K19945, and 18K15219.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Fukuhisa, H., Seki, N., Idichi, T. et al. Gene regulation by antitumor miR-130b-5p in pancreatic ductal adenocarcinoma: the clinical significance of oncogenic EPS8. J Hum Genet 64, 521–534 (2019). https://doi.org/10.1038/s10038-019-0584-6
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s10038-019-0584-6
This article is cited by
-
MicroRNA-130b Suppresses Malignant Behaviours and Inhibits the Activation of the PI3K/Akt Signaling Pathway by Targeting MET in Pancreatic Cancer
Biochemical Genetics (2025)
-
High-throughput analysis of ANRIL circRNA isoforms in human pancreatic islets
Scientific Reports (2022)
-
Noncoding RNA actions through IGFs and IGF binding proteins in cancer
Oncogene (2022)
-
Identification of ITPR1 as a Hub Gene of Group 3 Medulloblastoma and Coregulated Genes with Potential Prognostic Values
Journal of Molecular Neuroscience (2022)
-
Anillin is an emerging regulator of tumorigenesis, acting as a cortical cytoskeletal scaffold and a nuclear modulator of cancer cell differentiation
Cellular and Molecular Life Sciences (2021)