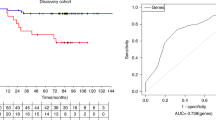
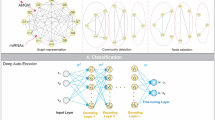
Abstract
Renal cell carcinoma is one of the most common urinary system tumors in adults, it is usually asymptomatic in its early stage and the patients are often diagnosed late. MicroRNA has a higher diagnostic accuracy than traditional markers and may become a new type of early diagnostic biomarker for kidney cancer. Three computational methods and several bioinformatic methods including PPI network, overall survival analysis and enrichment analysis were used to identify the significant differentially expressed miRNAs. Thirteen miRNAs that were significantly differentially expressed in RCC patients were identified, 10 of them have been proved to be associated with kidney cancer in other studies, miR-576, miR-616 and miR-133a-2 are three newly discovered biomarkers of RCC in this study. We found that the target genes of miR-576 (CUL3 and RAC1) are involved in the regulation of multiple cancer-related biological pathways, and the target gene of miR-616 (ASB13 and FBXW2) has been reported to be associated with the development of other cancers. Our findings may have guiding significance for the early diagnosis of renal cell carcinoma.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Cairns P. Renal cell carcinoma. Cancer Biomark. 2011;9:461.
Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2018. CA Cancer J Clin. 2018;68:7–30.
Butz H, Szabo PM, Nofech-Mozes R, Rotondo F, Kovacs K, Mirham L, et al. Integrative bioinformatics analysis reveals new prognostic biomarkers of clear cell renal cell carcinoma. Clin Chem. 2014;60:1314.
Sacco E, Pinto F, Sasso F, Racioppi M, Gulino G, Volpe A, et al. Paraneoplastic Syndromes in Patients with Urological Malignancies. Urol Int. 2009;83:1.
Flanigan RC, Campbell SC, Clark JI, Picken MM. Metastatic renal cell carcinoma. Curr Treat Option On. 2003;4:385–90.
Cohen HT, McGovern FJ. Renal-cell carcinoma. New Engl J Med. 2005;353:2477.
De Mulder PHM, van Herpen CML, Mulders PAF. Current treatment of renal cell carcinoma. Ann Oncol. 2004;15:v319.
Wasim AS, Mumtaz F. Limitations of CT scanning in Bosniak staging of renal cystic carcinoma. J Surg Case Rep. 2018;4:1–4.
Björklund M, Heikkilä P, Koivunen E. Peptide inhibition of catalytic and noncatalytic activities of matrix metalloproteinase-9 blocks tumor cell migration and invasion. J Biol Chem. 2004;279:29589.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97.
Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390.
Braicu C, Calin GA, Berindan-Neagoe I. MicroRNAs and cancer therapy - from bystanders to major players. Curr Med Chem. 2013;20:3561.
Faragalla H, Youssef YM, Scorilas A, Khalil B, White NMA, Mejia-Guerrero S, et al. The clinical utility of miR-21 as a diagnostic and prognostic marker for renal cell carcinoma. J Mol Diagn. 2012;14:385.
Gaudelot K, Gibier J, Pottier N, Hémon B, Van Seuningen I, Glowacki F, et al. Targeting miR-21 decreases expression of multi-drug resistant genes and promotes chemosensitivity of renal carcinoma. Tumor Biol. 2017;39:568834959.
Zhan C, Yan L, Wang L, Jiang W, Zhang Y, Xi J, et al. Identification of reference miRNAs in human tumors by TCGA miRNA-seq data. Biochem Bioph Res Co. 2014;453:375.
Wang H, Peng R, Wang J, Qin Z, Xue L. Circulating microRNAs as potential cancer biomarkers: the advantage and disadvantage. Clin Epigenetics. 2018;10:19.
Yoon D, Lee E, Park T. Robust imputation method for missing values in microarray data. BMC Bioinf. 2007;8(Suppl 2):S6.
Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, et al. Normalization for cDNA microarray data: a robust composite method addressing single andmultiple slide systematic variation. Nucleic Acids Res. 2002;30:e15.
Yuan L, Chen L, Qian K, Qian G, Wu C, Wang X. et al. Co-expression network analysis identified six hub genes in association with progression and prognosis in human clear cell renal cell carcinoma (ccRCC). Genomics Data. 2017;14:132.
Su Q, Wang Y, Jiang X, Chen F, Lu W. A cancer gene selection algorithm based on the K-S test and CFS. Biomed Res Int. 2017;2017:1.
Li S, Wang S, Guo Z, Wu H, Jin X, Wang Y, et al. miRNA profiling reveals dysregulation of RET and RET-regulating pathways in Hirschsprung’s Disease. PLoS ONE. 2016;11:e150222.
Yasmin F, Tun HM, Konya TB, Guttman DS, Chari RS, Field CJ, et al. Cesarean section, formula feeding, and infant antibiotic exposure: separate and combined impacts on gut microbial changes in later infancy. Front Pediatr. 2017;5:200.
Zhou Z, Cheng Y, Jiang Y, Liu S, Zhang M, Liu J, et al. Ten hub genes associated with progression and prognosis of pancreatic carcinoma identified by co-expression analysis. Int J Biol Sci. 2018;14:124.
Monteys AM, Spengler RM, Wan J, Tecedor L, Lennox KA, Xing Y, et al. Structure and activity of putative intronic miRNA promoters. RNA (New Y, N Y). 2010;16:495.
Juan D, Alexe G, Antes T, Liu H, Madabhushi A, Delisi C, et al. Identification of a microRNA panel for clear-cell kidney cancer. Urology. 2010;75:835.
Rasti A, Mehrazma M, Madjd Z, Keshtkar AA, Roudi R, Babashah S. Diagnostic and prognostic accuracy of miR-21 in renal cell carcinoma: a systematic review protocol. BMJ Open. 2016;6:e9667.
Ji H, Tian D, Zhang B, Zhang Y, Yan D, Wu S. Overexpression of miR-155 in clear-cell renal cell carcinoma and its oncogenic effect through targeting FOXO3a. Exp Ther Med. 2017;13:2286.
von Brandenstein M, Pandarakalam JJ, Kroon L, Loeser H, Herden J, Braun G, et al. MicroRNA 15a, inversely correlated to PKCα, is a potential marker to differentiate between benign and malignant renal tumors in biopsy and urine samples. Am J Pathol. 2012;180:1787.
Liu TY, Zhang H, Du SM, Li J, Wen XH. Expression of microRNA-210 in tissue and serum of renal carcinoma patients and its effect on renal carcinoma cell proliferation, apoptosis, and invasion. Genet Mol Res. 2016;15:15017746.
Hao JF, Ren KM, Bai JX, Wang SN, Shao B, Cao N, et al. Identification of potential biomarkers for clear cell renal cell carcinoma based on microRNA-mRNA pathway relationships. Special Issue 2. J Cancer Res Ther. 2014;10:C167–77.
Shi J, Zhuang Y, Liu XK, Zhang YX, Zhang Y. TGF-beta induced RBL2 expression in renal cancer cells by down-regulating miR-93. Clin Transl Oncol. 2014;16:986.
Prior C, Perez-Gracia JL, Garcia-Donas J, Rodriguez-Antona C, Guruceaga E, Esteban E, et al. Identification of tissue microRNAs predictive of sunitinib activity in patients with metastatic renal cell carcinoma. PLoS ONE. 2014;9:e86263.
Li W, Dou Z, We S, Zhu Z, Pan D, Jia Z, et al. Long noncoding RNA BDNF-AS is associated with clinical outcomes and has functional role in human prostate cancer. Biomed Pharm. 2018;102:1105.
Liu Z, Liu XW, Liu SA, Lv JJ, Fu Q. Clinical significance of changes of expression of the Wnt/beta-catenin signaling pathway in renal clear cell carcinoma. Eur Rev Med Pharm Sci. 2016;20:4840.
Gao Y, Li J, Qiao N, Meng Q, Zhang M, Wang X, et al. Adrenomedullin blockade suppresses sunitinib-resistant renal cell carcinoma growth by targeting the ERK/MAPK pathway. Oncotarget. 2016;7:63374.
Guo G, Gui Y, Gao S, Tang A, Hu X, Huang Y, et al. Frequent mutations of genes encoding ubiquitin-mediated proteolysis pathway components in clear cell renal cell carcinoma. Nat Genet. 2012;44:17.
Guo H, German P, Bai S, Barnes S, Guo W, Qi X, et al. The PI3K/AKT pathway and renal cell carcinoma. J Genet Genomics. 2015;42:343.
Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8:387.
Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13:11.
Lim W, Jeong W, Song G. Delphinidin suppresses proliferation and migration of human ovarian clear cell carcinoma cells through blocking AKT and ERK1/2 MAPK signaling pathways. Mol Cell Endocrinol. 2016;422:172.
Turney KD, Parrish AR, Orozco J, Gandolfi AJ. Selective activation in the MAPK pathway by Hg(II) in precision-cut rabbit renal cortical slices. Toxicol Appl Pharm. 1999;160:262.
Huang D, Ding Y, Luo WM, Bender S, Qian CN, Kort E, et al. Inhibition of MAPK kinase signaling pathways suppressed renal cell carcinoma growth and angiogenesis in vivo. Cancer Res. 2008;68:81.
Ciechanover A, Orian A, Schwartz AL. Ubiquitin-mediated proteolysis: biological regulation via destruction. Bioessays. 2000;22:442.
Vodermaier HC. APC/C and SCF: controlling each other and the cell cycle. Curr Biol. 2004;14:R787.
Blenk S, Engelmann J, Weniger M, Schultz J, Dittrich M, Rosenwald A, et al. Germinal center B cell-like (GCB) and activated B cell-like (ABC) type of diffuse large B cell lymphoma (DLBCL): analysis of molecular predictors, signatures, cell cycle state and patient survival. Cancer Inf. 2007;3:399.
Chi C, Murphy LC, Hu P. Recurrent copy number alterations in young women with breast cancer. Oncotarget. 2018;9:11541.
Yang F, Xu J, Li H, Tan M, Xiong X, Sun Y. FBXW2 suppresses migration and invasion of lung cancer cells via promoting β-catenin ubiquitylation and degradation. Nat Commun. 2019;10:1382.
Xu J, Zhou W, Yang F, Chen G, Li H, Zhao Y, et al. The β-TrCP-FBXW2-SKP2 axis regulates lung cancer cell growth with FBXW2 acting as a tumour suppressor. Nat Commun. 2017;8:14002.
Acknowledgements
This work was supported by the Beijing Municipal Education Commission (Grant No. KM201410005030), the Importation and Development of High-Caliber Talents Project of Beijing Municipal Institutions (Xiaoyi Zhang), the practical training plan of Beijing Municipal Education Commission (Xiaoyi Zhang), the National Natural Science Foundation of China (Grant No. 31100523).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tang, T., Du, X., Zhang, X. et al. Computational identification and analysis of early diagnostic biomarkers for kidney cancer. J Hum Genet 64, 1015–1022 (2019). https://doi.org/10.1038/s10038-019-0640-2
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s10038-019-0640-2