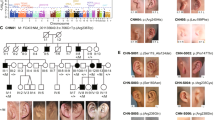
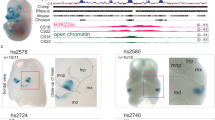
Abstract
Hemifacial microsomia (HFM) is a rare congenital disorder that affects facial symmetry, ear development, and other congenital anomalies. However, known causal genes account for only approximately 6% of patients, indicating the need to discover more pathogenic genes. Association tests demonstrated an association between common variants in SHROOM3 and HFM (P = 1.02E-4 for the lead SNP), while gene burden analysis revealed a significant enrichment of rare variants in HFM patients compared to healthy controls (P = 2.78E-5). We then evaluated the expression patterns of SHROOM3 and the consequences of its deleterious variants. Our study identified 7 deleterious variants in SHROOM3 among the 320 Chinese HFM patients and 2 deleterious variants in two HFM trios, respectively, suggesting a model of dominant inheritance with incomplete penetrance. These variants were predicted to significantly impact SHROOM3 function. Furthermore, the gene expression pattern of SHROOM3 in the pharyngeal arches and the presence of facial abnormalities in gene-edited mice suggest that SHROOM3 plays important roles in facial development. Our findings suggest that SHROOM3 is a likely pathogenic gene for HFM.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
All data are available upon request.
References
Luquetti DV, Cox TC, Lopez-Camelo J, Dutra MdaG, Cunningham ML, Castilla EE. Preferential associated anomalies in 818 cases of microtia in South America. Am J Med Genet A. 2013;161A:1051–7.
Luquetti DV, Heike CL, Hing AV, Cunningham ML, Cox TC. Microtia: epidemiology and genetics. Am J Med Genet A. 2012;158A:124–39.
Mao K, Borel C, Ansar M, Jolly A, Makrythanasis P, Froehlich C, et al. FOXI3 pathogenic variants cause one form of craniofacial microsomia. Nat Commun. 2023;14:2026.
Quiat D, Timberlake AT, Curran JJ, Cunningham ML, McDonough B, Artunduaga MA, et al. Damaging variants in FOXI3 cause microtia and craniofacial microsomia. Genet Med. 2023;25:143–50.
Timberlake AT, Griffin C, Heike CL, Hing AV, Cunningham ML, Chitayat D, et al. Haploinsufficiency of SF3B2 causes craniofacial microsomia. Nat Commun. 2021;12:4680.
Luquetti DV, Heike CL, Zarante I, Timms AE, Gustafson J, Pachajoa H, et al. MYT1 role in the microtia-craniofacial microsomia spectrum. Mol Genet Genomic Med. 2020;8:e1401.
Quiat D, Kim SW, Zhang Q, Morton SU, Pereira AC, DePalma SR, et al. An ancient founder mutation located between ROBO1 and ROBO2 is responsible for increased microtia risk in Amerindigenous populations. Proc Natl Acad Sci USA. 2022;119:e2203928119.
Zhang YB, Hu J, Zhang J, Zhou X, Li X, Gu C, et al. Genome-wide association study identifies multiple susceptibility loci for craniofacial microsomia. Nat Commun. 2016;7:10605.
Xu X, Wang B, Jiang Z, Chen Q, Mao K, Shi X, et al. Novel risk factors for craniofacial microsomia and assessment of their utility in clinic diagnosis. Hum Mol Genet. 2021;30:1045–56.
Ernst S, Liu K, Agarwala S, Moratscheck N, Avci ME, Dalle Nogare D, et al. Shroom3 is required downstream of FGF signalling to mediate proneuromast assembly in zebrafish. Development. 2012;139:4571–81.
Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–6.
Ray D, Venkataraghavan S, Zhang W, Leslie EJ, Hetmanski JB, Weinberg SM, et al. Pleiotropy method reveals genetic overlap between orofacial clefts at multiple novel loci from GWAS of multi-ethnic trios. PLoS Genet. 2021;17:e1009584.
Leslie EJ, Carlson JC, Shaffer JR, Butali A, Buxo CJ, Castilla EE, et al. Genome-wide meta-analyses of nonsyndromic orofacial clefts identify novel associations between FOXE1 and all orofacial clefts, and TP63 and cleft lip with or without cleft palate. Hum Genet. 2017;136:275–86.
Diaz Perez KK, Chung S, Head ST, Epstein MP, Hecht JT, Wehby GL, et al. Rare variants found in multiplex families with orofacial clefts: does expanding the phenotype make a difference? Am J Med Genet A. 2023;191:2558–70.
Chen Z, Kuang L, Finnell RH, Wang H. Genetic and functional analysis of SHROOM1-4 in a Chinese neural tube defect cohort. Hum Genet. 2018;137:195–202.
Lemay P, Guyot MC, Tremblay E, Dionne-Laporte A, Spiegelman D, Henrion E, et al. Loss-of-function de novo mutations play an important role in severe human neural tube defects. J Med Genet. 2015;52:493–7.
Xu L, Yang K, Yin S, Gu Y, Fan Q, Wang Y, et al. Family-based exome sequencing identifies candidate genes related to keratoconus in Chinese families. Front Genet. 2022;13:988620.
Hildebrand JD, Soriano P. Shroom, a PDZ domain-containing actin-binding protein, is required for neural tube morphogenesis in mice. Cell. 1999;99:485–97.
Cong PK, Bai WY, Li JC, Yang MY, Khederzadeh S, Gai SR, et al. Genomic analyses of 10,376 individuals in the Westlake BioBank for Chinese (WBBC) pilot project. Nat Commun. 2022;13:2939.
Xu Y, Zhang T, Zhou Q, Hu M, Qi Y, Xue Y, et al. A single-cell transcriptome atlas profiles early organogenesis in human embryos. Nat Cell Biol. 2023;25:604–15.
Diez-Roux G, Banfi S, Sultan M, Geffers L, Anand S, Rozado D, et al. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 2011;9:e1000582.
Prokop JW, Yeo NC, Ottmann C, Chhetri SB, Florus KL, Ross EJ, et al. Characterization of coding/noncoding variants for SHROOM3 in patients with CKD. J Am Soc Nephrol. 2018;29:1525–35.
Yeo NC, O’Meara CC, Bonomo JA, Veth KN, Tomar R, Flister MJ, et al. Shroom3 contributes to the maintenance of the glomerular filtration barrier integrity. Genome Res. 2015;25:57–65.
Nishimura T, Takeichi M. Shroom3-mediated recruitment of Rho kinases to the apical cell junctions regulates epithelial and neuroepithelial planar remodeling. Development. 2008;135:1493–502.
Plageman TF Jr, Chung MI, Lou M, Smith AN, Hildebrand JD, Wallingford JB, et al. Pax6-dependent Shroom3 expression regulates apical constriction during lens placode invagination. Development. 2010;137:405–15.
Plageman TF Jr, Chauhan BK, Yang C, Jaudon F, Shang X, Zheng Y, et al. A Trio-RhoA-Shroom3 pathway is required for apical constriction and epithelial invagination. Development. 2011;138:5177–88.
Carlson BM. Human embryology and developmental biology. Sixth edition. ed. Elsevier: St. Louis, Missouri, 2019.
Schlosser G. Induction and specification of cranial placodes. Dev Biol. 2006;294:303–51.
Rossant J, Tam PPL. ScienceDirect. Mouse development: patterning, morphogenesis, and organogenesis. Academic Press: San Diego, 2002.
Lewandowski JP, Du F, Zhang S, Powell MB, Falkenstein KN, Ji H, et al. Spatiotemporal regulation of GLI target genes in the mammalian limb bud. Dev Biol. 2015;406:92–103.
Groves AK, Fekete DM. Shaping sound in space: the regulation of inner ear patterning. Development. 2012;139:245–57.
Lee HJ, Zheng JJ. PDZ domains and their binding partners: structure, specificity, and modification. Cell Commun Signal. 2010;8:8.
Kim K, Ossipova O, Sokol SY. Neural crest specification by inhibition of the ROCK/Myosin II pathway. Stem Cells. 2015;33:674–85.
Dietz ML, Bernaciak TM, Vendetti F, Kielec JM, Hildebrand JD. Differential actin-dependent localization modulates the evolutionarily conserved activity of Shroom family proteins. J Biol Chem. 2006;281:20542–54.
Acknowledgements
We thank the patients and their guardians for their participation in this research. This work was supported by the National Natural Science Foundation of China (81701930 to B.Q.W.).
Author information
Authors and Affiliations
Contributions
Conceptualization: B.W., W.Z.; Sample collection: W.Z., B.-H.Z., P.L.; Experiments: Q.L., B.-H.Z., Q.C.; Data analysis: Q.L., B.-H.Z., X.Z.; Writing-original draft: B.W., Q.L., W.Z.; Writing-reviewing and editing: W.Z., B.W.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflicts of interest.
Ethics
All human studies were reviewed and approved by the Ethics Committee of the Eye & ENT Hospital of Fudan University. An informed consent form was signed by all individuals, and explicit written consent was provided for publication of clinical images.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, Q., Zhang, BH., Chen, Q. et al. Pathogenic variants in SHROOM3 associated with hemifacial microsomia. J Hum Genet 70, 189–194 (2025). https://doi.org/10.1038/s10038-025-01317-1
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s10038-025-01317-1