Abstract
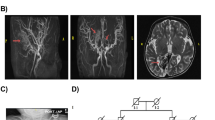
Cobalamin (Cbl) metabolism deficiencies are a heterogeneous group (CblA, CblB, CblC, CblD, CblE, CblF, CblG) of autosomal recessive disorders. CblF deficiency occurs due to mutations in LMBRD1 gene, causing variable phenotype, including neurological, haematological, developmental and dermatological defects. Here, we describe a 15-year-old male, presented with clinical features of Dyskeratosis Congenita (DC) such as dystrophic nails, skin discoloration with additional clinical features of uniform reticulate-brown hued hyperpigmentation, developmental delay, mild intellectual disability, mucositis and anemia. Genomic analysis using high throughput next generation sequencing (NGS) identified a novel splice site deletion (c.562+4_562+7del) in the LMBRD1 gene resulting in Cbl deficiency. cDNA sequencing elucidated exon 6 skipping as a consequence of a novel deletion, resulting in significant structural alterations of LMBD1 protein, which was further validated by in-silico computational analysis. Computational modeling and docking studies revealed a reduced interaction affinity between the LMBD1 protein and its partner protein ABCD4. These alterations contribute to a disrupted cascade mechanism in cobalamin (Cbl) metabolism resulting in development of variable clinical phenotypes. In our case, the proband was treated with intravenous hydroxocobalamin therapy and follow up showed a significant improvement in clinical symptoms of skin hyperpigmentation, angular cheilitis and aphthous ulcers. Hence the genomic analysis is essentially important for the appropriate genetic counseling and management of the disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The novel variant identified in the present study has been submitted in ClinVar database under the submission ID SUB13687835 (https://submit.ncbi.nlm.nih.gov/subs/clinvar_file/SUB13687835/overview) (ClinVar accession ID: VCV002572168.1).
References
Wahbeh F, Manyama M. The role of Vitamin B12 and genetic risk factors in the etiology of neural tube defects: A systematic review. Int J Developmental Neurosci. 2021;81(5):386–406.
Rosenblatt DS, Cooper BA. Inherited disorders of vitamin B12 utilization. Bioessays. 1990;12(7):331–4.
Ross AC, Caballero B, Cousins RJ, Tucker KL Modern nutrition in health and disease. Jones & Bartlett Learning; 2020.
Rosenblatt DS, Hosack A, Matiaszuk NV, Cooper BA, Laframboise R. Defect in vitamin B12 release from lysosomes: newly described inborn error of vitamin B12 metabolism. Science. 1985;228(4705):1319–21.
Shih VE, Axel SM, Tewksbury JC, Watkins D, Cooper BA, Rosenblatt DS. Defective lysosomal release of vitamin B12 (cblF): a hereditary cobalamin metabolic disorder associated with sudden death. Am J Med Genet. 1989;33(4):555–63.
MacDonald MR, Wiltse HE, Bever JL, Rosenblatt DS. Clinical heterogeneity in two patients with cblF disease. Am J Hum Genet. 1992;51(suppl):A353.
Waggoner DJ, Ueda K, Mantia C, Dowton SB. Methylmalonic aciduria (cblF): case report and response to therapy. Am J Med Genet. 1998;79(5):373–5.
Rutsch F, Gailus S, Miousse IR, Suormala T, Sagné C, Toliat MR, et al. Identification of a putative lysosomal cobalamin exporter altered in the cblF defect of vitamin B12 metabolism. Nat Genet. 2009;41(2):234–9.
Gailus S, Suormala T, Malerczyk-Aktas AG, Toliat MR, Wittkampf T, Stucki M, et al. A novel mutation in LMBRD1 causes the cblF defect of vitamin B 12 metabolism in a Turkish patient. J Inherit Metab Dis. 2010;33:17–24.
Miousse IR, Watkins D, Rosenblatt DS. Novel splice site mutations and a large deletion in three patients with the cblF inborn error of vitamin B12 metabolism. Mol Genet Metab. 2011;102(4):505–7.
Rutsch F, Gailus S, Suormala T, Fowler B. LMBRD1: the gene for the cblF defect of vitamin B 12 metabolism. J Inherit Metab Dis. 2011;34:121–6.
Farwell Gonzalez KD, Li X, Lu HM, Lu H, Pellegrino JE, Miller RT, et al. Diagnostic exome sequencing and tailored bioinformatics of the parents of a deceased child with cobalamin deficiency suggests digenic inheritance of the MTR and LMBRD1 genes. JIMD Rep. 2015;15:29–37.
Constantinou P, D’Alessandro M, Lochhead P, Samant S, Bisset WM, Hauptfleisch C, et al. Deciphering developmental disorders study group. a new, atypical case of cobalamin F disorder diagnosed by whole exome sequencing. Mol Syndromol. 2016;6(5):254–8.
Tong F, Yang RL, Chen R, Zhao ZY. Case report the first case of cobalamin F disorder in China: report and literature review. HK J Paediatr (N. Ser). 2019;24(1):43–7.
Altawil L, Alshihry H, Ahmed H, Shamseldin HE, Alkuraya F. Vitamin B12 deficiency secondary to cobalamin F deficiency simulating dyskeratosis congenita. JAAD Case Rep. 2020;6(9):882–5.
Braz SV, Benicio RA, Tonelli GS, Báo SN, Moretti PN, Pic‐Taylor A, et al. Cobalamin F deficiency in a girl with severe skin hyperpigmentation and a homozygous LMBRD1 variant. Clin Exp Dermatol. 2022;47(4):812–5.
Kitai K, Kawaguchi K, Tomohiro T, Morita M, So T, Imanaka T. The lysosomal protein ABCD4 can transport vitamin B12 across liposomal membranes in vitro. J Biolog Chem. 2021;296:100654.
Sun CY, Chang SC, Wang HP, Lee YJ, Pan KH, Lin CL, et al. LMBD1 protein participates in cell mitosis by regulating microtubule assembly. Biochemical J. 2021;478(12):2321–37.
Mucha P, Kus F, Cysewski D, Smolenski RT, Tomczyk M. Vitamin B12 metabolism: a network of multi-protein mediated processes. Int J Mol Sci. 2024;25(15):8021.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–23.
Lobley A, Sadowski MI, Jones DT. pGenTHREADER and pDomTHREADER: new methods for improved protein fold recognition and superfamily discrimination. Bioinformatics. 2009;25(14):1761–7.
Šali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234(3):779–815.
Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26(16):1701–18.
Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26(2):283–91.
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The protein data bank. Nucleic Acids Res. 2000;28(1):235–42.
Xu D, Feng Z, Hou WT, Jiang YL, Wang L, Sun L, et al. Cryo-EM structure of human lysosomal cobalamin exporter ABCD4. Cell Res. 2019;29(12):1039–41.
Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein− protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125(7):1731–7.
Xue LC, Rodrigues JP, Kastritis PL, Bonvin AM, Vangone A. PRODIGY: a web server for predicting the binding affinity of protein–protein complexes. Bioinformatics. 2016;32(23):3676–8.
Laskowski RA. PDBsum: summaries and analyses of PDB structures. Nucleic acids Res. 2001;29(1):221–2.
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J computational Chem. 2004;25(13):1605–12.
Ulak M, Kvestad I, Chandyo RK, Schwinger C, Basnet S, Shrestha M, et al. The effect of vitamin b12 supplementation on leukocyte telomere length in mildly stunted nepalese children: a secondary outcome of a randomized controlled trial. J Nutr. 2024;154(8):2543–50.
Deme JC, Hancock MA, Xia X, Shintre CA, Plesa M, Kim JC, et al. Purification and interaction analyses of two human lysosomal vitamin B12 transporters: LMBD1 and ABCD4. Mol Membr Biol. 2014;31(7-8):250–61.
Acknowledgements
This work was supported by ICMR-National Institute of Immunohaematology. We thank resident doctors from hematology department of KEM hospital for clinical diagnosis and treatment of subject. We thank the subject and his family for their cooperation and participation in the study. Thanks also due to Council of Scientific and Industrial Research for providing fellowship to AS.
Funding
This study was performed with the financial support by the intramural grant from ICMR-National Institute of Immunohaematology, India under grant no. [ICMR/NIIH/15-2019]. The research was supported by fellowship grant by Council of Scientific and Industrial Research (CSIR) NET-JRF awarded to AS. The funding body played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
AS collected clinical history and performed laboratory work. CS and SK carried out clinical diagnosis, treatment and follow up. AS and SK has equally contributed. AS and MG performed molecular work, genetic analysis along with family screening and follow up. AS, SK and BRV contributed to conception and designing of the work. SKC performed sequence and structure based computational analysis of mutant LMBD1 protein. AS, SK, SKC reviewed a literature and drafted a manuscript. BRV and CS performed general supervision of the overall research work. AS deposited variant on ClinVar database. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no competing interests.
Ethical approval and consent participation
The study was approved by the “Institutional Ethics Committee on human subjects” of ICMR-National Institute of Immunohaematology (Ethics approval number: NIIH/IEC/22-2019). All procedures performed in studies involving human participants were as per the ethical standards of the institutional review board of the institute and with the 1964 declaration of Helsinki (DoH) and its later amendments or comparable ethical standards.
Consent for publication
Written informed consent was obtained from the participant’s mother to use images and other personal or clinical details.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shah, A., Khuba, S., Kumar C, S. et al. Phenotype puzzle: the role of novel LMBRD1 gene variant in Cbl deficiency causing Dyskeratosis Congenita-like clinical manifestations. J Hum Genet 70, 207–213 (2025). https://doi.org/10.1038/s10038-025-01320-6
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s10038-025-01320-6