Abstract
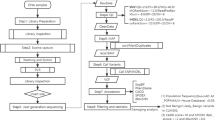
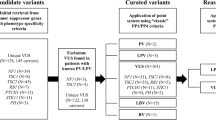
Although whole-exome sequencing (WES) is now widely used to diagnose neonatal genetic diseases, the genetic causes in over half of the cases remain unresolved, primarily due to variants of uncertain significance (VUS). Therefore, reclassifying VUS may be an effective strategy to improve WES’s diagnostic yield. However, not all reclassification approaches are suitable for clinicians. Patients in the neonatal unit of Hebei Provincial Children’s Hospital who underwent WES for suspected genetic diseases and demonstrated VUS were re-evaluated from January 2019 to December 2023 using user-friendly methods. A total of 676 individuals were tested, with 101 phenotype-associated VUS identified in 82 patients. Thirty (29.7%) VUS classifications were changed: 24 were upgraded to likely pathogenic or pathogenic, and 6 were downgraded to likely benign. VUS reclassification clarified the molecular diagnosis in 19 cases, increasing the WES diagnostic rate from 30.2% to 33.0%. Computational prediction contributed the most to reclassification, whereas clinical phenotype-related evidence was also particularly significant in upgrading variants. Moreover, phenotype-associated VUS with a score of ≥3 points are more likely to be classified as likely pathogenic or pathogenic, thus requiring more attention. This study provides a practical reference for clinicians in managing VUS reclassification.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
References
Yang L, Liu X, Li Z, Zhang P, Wu B, Wang H, et al. Genetic aetiology of early infant deaths in a neonatal intensive care unit. J Med Genet. 2020;57:169–77.
Meng L, Pammi M, Saronwala A, Magoulas P, Ghazi AR, Vetrini F, et al. Use of exome sequencing for infants in intensive care units: ascertainment of severe single-gene disorders and effect on medical management. JAMA Pediatr. 2017;171:e173438.
Tan TY, Dillon OJ, Stark Z, Schofield D, Alam K, Shrestha R, et al. Diagnostic impact and cost-effectiveness of whole-exome sequencing for ambulant children with suspected monogenic conditions. JAMA Pediatr. 2017;171:855–62.
Wright CF, FitzPatrick DR, Firth HV. Paediatric genomics: diagnosing rare disease in children. Nat Rev Genet. 2018;19:253–68.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Clift K, Macklin S, Halverson C, McCormick JB, Abu Dabrh AM, Hines S. Patients’ views on variants of uncertain significance across indications. J Community Genet. 2020;11:139–45.
Tsai GJ, Rañola JMO, Smith C, Garrett LT, Bergquist T, Casadei S, et al. Outcomes of 92 patient-driven family studies for reclassification of variants of uncertain significance. Genet Med. 2019;21:1435–42.
SoRelle JA, Thodeson DM, Arnold S, Gotway G, Park JY. Clinical utility of reinterpreting previously reported genomic epilepsy test results for pediatric patients. JAMA Pediatr. 2019;173:e182302.
So M-K, Jeong T-D, Lim W, Moon B-I, Paik NS, Kim SC, et al. Reinterpretation of BRCA1 and BRCA2 variants of uncertain significance in patients with hereditary breast/ovarian cancer using the ACMG/AMP 2015 guidelines. Breast Cancer. 2019;26:510–9.
Bennett JS, Bernhardt M, McBride KL, Reshmi SC, Zmuda E, Kertesz NJ, et al. Reclassification of variants of uncertain significance in children with inherited arrhythmia syndromes is predicted by clinical factors. Pediatr Cardiol. 2019;40:1679–87.
Wright M, Menon V, Taylor L, Shashidharan M, Westercamp T, Ternent CA. Factors predicting reclassification of variants of unknown significance. Am J Surg. 2018;216:1148–54.
Chen E, Facio FM, Aradhya KW, Rojahn S, Hatchell KE, Aguilar S, et al. Rates and classification of variants of uncertain significance in hereditary disease genetic testing. JAMA Netw Open. 2023;6:e2339571.
Chourasia N, Vaidya R, Sengupta S, Mefford HC, Wheless J. A Retrospective review of reclassification of variants of uncertain significance in a pediatric epilepsy cohort undergoing genetic panel testing. Pediatr Neurol. 2024;161:101–7.
Zhang L, Shen M, Shu X, Zhou J, Ding J, Lin H, et al. The recommendation of re-classification of variants of uncertain significance (VUS) in adult genetic disorders patients. J Hum Genet. 2024;69:425–31.
Fowler DM, Rehm HL. Will variants of uncertain significance still exist in 2030? Am J Hum Genet. 2024;111:5–10.
Tavtigian SV, Harrison SM, Boucher KM, Biesecker LG. Fitting a naturally scaled point system to the ACMG/AMP variant classification guidelines. Hum Mutat. 2020;41:1734–7.
Ellard S, Baple EL, Callaway A, Berry I, Forrester N, Turnbull C, et al. ACGS best practice guidelines for variant classification in rare disease 2020. http://www.acgs.uk.com/quality/best-practice-guidelines/.
Sequence Variant Interpretation, ClinGen. https://clinicalgenome.org/working-groups/sequence-variant-interpretation/.
Zhang J, Yao Y, He H, Shen J. Clinical interpretation of sequence variants. Curr Protoc Hum Genet. 2020;106:e98.
Rehm HL, Bale SJ, Bayrak-Toydemir P, Berg JS, Brown KK, Deignan JL, et al. ACMG clinical laboratory standards for next-generation sequencing. Genet Med. 2013;15:733–47.
Zhu Y, Cao Y, Ma L, Fan L, Pu W, Xia Y. Trichohepatoenteric syndrome and cytomegalovirus infection: case report and literature summary. SAGE Open Med Case Rep. 2024;12:2050313X241248393.
Ackerman MJ. Genetic purgatory and the cardiac channelopathies: exposing the variants of uncertain/unknown significance issue. Heart Rhythm. 2015;12:2325–31.
Mercati O, Abi Warde M-T, Lina-Granade G, Rio M, Heide S, de Lonlay P, et al. PRPS1 loss-of-function variants, from isolated hearing loss to severe congenital encephalopathy: new cases and literature review. Eur J Med Genet. 2020;63:104033.
Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, et al. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet. 2016;99:877–85.
Pejaver V, Byrne AB, Feng B-J, Pagel KA, Mooney SD, Karchin R, et al. Calibration of computational tools for missense variant pathogenicity classification and ClinGen recommendations for PP3/BP4 criteria. Am J Hum Genet. 2022;109:2163–77.
He W-B, Xiao W-J, Dai C-L, Wang Y-R, Li X-R, Gong F, et al. RNA splicing analysis contributes to reclassifying variants of uncertain significance and improves the diagnosis of monogenic disorders. J Med Genet. 2022;59:1010–6.
Wenger AM, Guturu H, Bernstein JA, Bejerano G. Systematic reanalysis of clinical exome data yields additional diagnoses: implications for providers. Genet Med. 2017;19:209–14.
Macklin SK, Jackson JL, Atwal PS, Hines SL. Physician interpretation of variants of uncertain significance. Fam Cancer. 2019;18:121–6.
Cherny S, Olson R, Chiodo K, Balmert LC, Webster G. Changes in genetic variant results over time in pediatric cardiomyopathy and electrophysiology. J Genet Couns. 2021;30:229–36.
David KL, Best RG, Brenman LM, Bush L, Deignan JL, Flannery D, et al. Patient re-contact after revision of genomic test results: points to consider—a statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2019;21:769–71.
Acknowledgements
We thank the patients and their parents for their cooperation in this study, and Editage (www.Editage.cn) for English language editing.
Funding
This study was supported by the Government Funded Clinical Medicine Excellent Talent Training Project (ZF2022034).
Author information
Authors and Affiliations
Contributions
XW wrote the main manuscript text. JJ, WP, YX, XY, and WG contributed to patient follow-up and data collection. LM and YC initiated and oversaw the research effort, writing, and other aspects of this project. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The studies involving human participants were reviewed and approved by the Ethics Committee of Children’s Hospital of Hebei Province (202457), and the requirement for informed consent was waived due to the retrospective nature of the study.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wu, X., Jiao, J., Pu, W. et al. Reclassification of variants of uncertain significance in neonatal genetic diseases: implications from a clinician’s perspective. J Hum Genet 70, 405–413 (2025). https://doi.org/10.1038/s10038-025-01348-8
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s10038-025-01348-8