Abstract
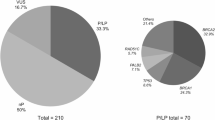
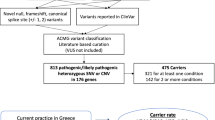
Secondary findings (SF) are pathogenic or likely pathogenic variants in genes unrelated to the primary purpose of genetic testing. The American College of Medical Genetics (ACMG) provides guidelines on which SF should be reported, involving 81 genes linked to different conditions. With the increasing use of genome sequencing (GS), SF are more frequently detected, presenting challenges for healthcare systems. The Brazilian population is often underrepresented in genomic studies, which limits population-specific knowledge. This study aimed to outline the profile of SF in the Brazilian Rare Genomes Project (BRGP). We analyzed retrospectively SF (ACMG) data from GS of 5402 BRGP individuals. Of the 5316 cases who consented to receive SF, 3.6% (191 cases) had at least one SF. The most common genes identified were TTR, TTN, and BRCA2. SF were mainly related to cardiovascular conditions (40.2%) and cancer predisposition (37.6%). Some variants, such as TTR: c.424G>A; p. (Val142Ile) and TP53: c.1010G>A; p. (Arg337His), were recurrent, reflecting population-specific traits and founder effects. Novel variants were 10.6% of SF. SF rate varies across studies and populations. While SF can aid early diagnosis, their relevance is debated due to potential psychological and healthcare burdens. Effective genetic counseling and public health policies are essential.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–74. https://doi.org/10.1038/gim.2013.73.
Miller DT, Lee K, Abul-Husn NS, Amendola LM, Brothers K, Chung WK, et al. ACMG SF v3.2 list for reporting of secondary findings in clinical exome and genome sequencing: a policy statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2023;25:100866. https://doi.org/10.1016/j.gim.2023.100866.
Elfatih A, Saad C, The Qatar Genome Program Research Consortium, Qatar Genome Project Management, Ismail S, Al-Muftah W, et al. Analysis of 14,392 whole genomes reveals 3.5% of Qataris carry medically actionable variants. Eur J Hum Genet. 2024;32:1465–73. https://doi.org/10.1038/s41431-024-01656-1.
Gordon AS, Zouk H, Venner E, Eng CM, Funke BH, Amendola LM, et al. Frequency of genomic secondary findings among 21,915 eMERGE network participants. Genet Med. 2020;22:1470–7. https://doi.org/10.1038/s41436-020-0810-9.
Saeidian AH, March ME, Youssefian L, Watson DJ, Bhandari E, Wang X, et al. Secondary ACMG and non-ACMG genetic findings in a multiethnic cohort of 16,713 pediatric participants. Genet Med. 2024;26:101225. https://doi.org/10.1016/j.gim.2024.101225.
Quaio CRDC, Moreira CM, Novo-Filho GM, Sacramento-Bobotis PR, Groenner Penna M, Perazzio SF, et al. Diagnostic power and clinical impact of exome sequencing in a cohort of 500 patients with rare diseases. Am J Med Genet C Semin Med Genet. 2020;184:955–64. https://doi.org/10.1002/ajmg.c.31860.
Naslavsky MS, Yamamoto GL, Almeida TF, Ezquina SAM, Sunaga DY, Pho N, et al. Exomic variants of an elderly cohort of Brazilians in the ABraOM database. Hum Mutat. 2017;38:751–63. https://doi.org/10.1002/humu.23220.
Naslavsky MS, Scliar MO, Yamamoto GL, Wang JYT, Zverinova S, Karp T, et al. Whole-genome sequencing of 1,171 elderly admixed individuals from Brazil. Nat Commun. 2022;13. https://doi.org/10.1038/s41467-022-28648-3
Coelho AVC, Mascaro-Cordeiro B, Lucon DR, Nóbrega MS, Reis RS, de Alexandre RB, et al. The Brazilian Rare Genomes Project: Validation of whole genome sequencing for rare diseases diagnosis. Front Mol Biosci. 2022;9. https://doi.org/10.3389/fmolb.2022.821582
den Dunnen JT, Dalgleish R, Maglott DR, Hart RK, Greenblatt MS, McGowan-Jordan J, et al. HGVS recommendations for the description of sequence variants: 2016 update. Hum Mutat. 2016;37:564–9. https://doi.org/10.1002/humu.22981.
McGowan-Jordan J, Hastings RJ, Moore S. ISCN 2020: an international system for human cytogenomic nomenclature. Cytogenetic Genome Res. 2020:160;7–8.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. https://doi.org/10.1038/gim.2015.30.
Riggs ER, Andersen EF, Cherry AM, Kantarci S, Kearney H, Patel A, et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen. Genet Med. 2020;22:245–57. https://doi.org/10.1038/s41436-019-0686-8.
Kalia SS, Adelman K, Bale SJ, Chung WK, Eng C, Evans JP, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19:249–55. https://doi.org/10.1038/gim.2016.190.
Miller DT, Lee K, Chung WK, Gordon AS, Herman GE, Klein TE, et al. ACMG SF v3.0 list for reporting of secondary findings in clinical exome and genome sequencing: a policy statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2021;23:1381–90. https://doi.org/10.1038/s41436-021-01172-3.
Miller DT, Lee K, Abul-Husn NS, Amendola LM, Brothers K, Chung WK, et al. ACMG SF v3.1 list for reporting of secondary findings in clinical exome and genome sequencing: a policy statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2022;24:1407–14. https://doi.org/10.1016/j.gim.2022.04.006.
Nolan J, Buchanan J, Taylor J, Almeida J, Bedenham T, Blair E, et al. Secondary (additional) findings from the 100,000 Genomes Project: disease manifestation, health care outcomes, and costs of disclosure. Genet Med. 2024;26:101051. https://doi.org/10.1016/j.gim.2023.101051.
Kim Y, Kim J-M, Cho H-W, Park H-Y, Park M-H. Frequency of actionable secondary findings in 7472 Korean genomes derived from the National Project of Bio Big Data pilot study. Hum Genet. 2023;142:1561–9. https://doi.org/10.1007/s00439-023-02592-8.
Aloraini T, Alsubaie L, Alasker S, Al Muitiri A, Alswaid A, Eyiad W, et al. The rate of secondary genomic findings in the Saudi population. Am J Med Genet A. 2022;188:83–88. https://doi.org/10.1002/ajmg.a.62491.
Johnston JJ, Brennan M-L, Radenbaugh B, Yoo SJ, Hernandez SM, NHGRI Reverse Phenotyping Core, et al. The ACMG SF v3.0 gene list increases returnable variant detection by 22% when compared with v2.0 in the ClinSeq cohort. Genet Med. 2022;24:736–43. https://doi.org/10.1016/j.gim.2021.11.012.
Yamashita T, Hamidi Asl K, Yazaki M, Benson MD. A prospective evaluation of the transthyretin Ile122 allele frequency in an African-American population. Amyloid. 2005;12:127–30. https://doi.org/10.1080/13506120500107162.
Censo 2022. Gov.br. 2022. https://www.ibge.gov.br/estatisticas/sociais/trabalho/22827-censo-demografico-2022.html. Accessed 28 Jan2025.
Skrahin A, Cheema HA, Hussain M, Rana NN, Rehman KU, Kumar R, et al. Secondary findings in a large Pakistani cohort tested with whole genome sequencing. Life Sci Alliance. 2023;6:e202201673. https://doi.org/10.26508/lsa.202201673.
Garritano S, Gemignani F, Palmero EI, Olivier M, Martel-Planche G, Le Calvez-Kelm F, et al. Detailed haplotype analysis at the TP53 locus in p.R337H mutation carriers in the population of Southern Brazil: evidence for a founder effect. Hum Mutat. 2010;31:143–50. https://doi.org/10.1002/humu.21151.
Corrêa TS, Asprino PF, de Oliveira ESC, Leite ACR, Weis L, Achatz MI, et al. TP53 p.R337H germline variant among women at risk of hereditary breast cancer in a public health system of Midwest Brazil. Genes. 2024;15:928 https://doi.org/10.3390/genes15070928.
Hunter JE, Jenkins CL, Bulkley JE, Gilmore MJ, Lee K, Pak CM, et al. ClinGen’s Pediatric Actionability Working Group: clinical actionability of secondary findings from genome-scale sequencing in children and adolescents. Genet Med. 2022;24:1328–35. https://doi.org/10.1016/j.gim.2022.02.019.
Virgens CSDAS. Variantes genéticas nos genes BRCA1 e BRCA2 em uma população da Bahia. Ufba.br. 2023. https://biologia.ufba.br/sites/biologia.ufba.br/files/tcc_final_-_cleiton_santos_das_virgens_.pdf. Accessed 28 Jan 2025.
Palmero EI, Carraro DM, Alemar B, Moreira MAM, Ribeiro-Dos-Santos Â, Abe-Sandes K, et al. The germline mutational landscape of BRCA1 and BRCA2 in Brazil. Sci Rep. 2018;8:9188. https://doi.org/10.1038/s41598-018-27315-2.
Mazzonetto P, Milanezi F, D’Andrea M, Martins S, Monfredini PM, Dos Santos Silva J, et al. BRCA1 and BRCA2 germline mutation analysis from a cohort of 1267 patients at high risk for breast cancer in Brazil. Breast Cancer Res Treat. 2023;199:127–36. https://doi.org/10.1007/s10549-023-06892-5.
de Oliveira JM, Zurro NB, Coelho AVC, Caraciolo MP, de Alexandre RB, Cervato MC, et al. The genetics of hereditary cancer risk syndromes in Brazil: a comprehensive analysis of 1682 patients. Eur J Hum Genet. 2022;30:818–23. https://doi.org/10.1038/s41431-022-01098-7.
Rodríguez-Salgado LE, Silva-Aldana CT, Medina-Méndez E, Bareño-Silva J, Arcos-Burgos M, Silgado-Guzmán DF, et al. Frequency of actionable Exomic secondary findings in 160 Colombian patients: Impact in the healthcare system. Gene. 2022;838:146699. https://doi.org/10.1016/j.gene.2022.146699.
Aarabi M, Darabi H, Bashar A, Bellissimo D, Rajkovic A, Yatsenko SA. Copy-number variants in the ACMG secondary finding genes: a reporting framework for clinical cytogeneticists. Genet Med Open. 2024;2:101839. https://doi.org/10.1016/j.gimo.2024.101839.
Yatsenko SA, Aarabi M, Hu J, Surti U, Ortiz D, Madan-Khetarpal S, et al. Copy number alterations involving 59 ACMG-recommended secondary findings genes. Clin Genet. 2020;98:577–88. https://doi.org/10.1111/cge.13852.
Acknowledgements
We would like to express our gratitude to the participants of this study and their families. This research was made possible by the data and findings provided by the Rare Genomes Project, an initiative of the Hospital Israelita Albert Einstein (HIAE) in collaboration with the Programa de Apoio ao Desenvolvimento Institucional do Sistema Único de Saúde (PROADI-SUS) of the Brazilian Ministry of Health (Law 12.101/2009). We also extend our thanks to the reference participant centers involved in this project.
Author information
Authors and Affiliations
Contributions
EP and LV contributed to conceptualization, data curation, formal analysis, investigation, methodology, validation, visualization, writing - original draft, and writing - review & editing; AVCC, MF, CAM, JRMP, JGAE, MM, TYTS, CRACQ, JRMC, KC, RMM, ACBT, RYY, VPC, LSS, GPC, RMRS contributed to writing - review & editing; KOP, JBOF e TFA contributed to project administration and funding acquisition. All authors approved the final draft.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Perrone, E., Virmond, L., Coelho, A.V.C. et al. ACMG secondary findings in the Brazilian rare genomes project: insights from 5402 genome sequencing. J Hum Genet 70, 415–420 (2025). https://doi.org/10.1038/s10038-025-01349-7
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s10038-025-01349-7