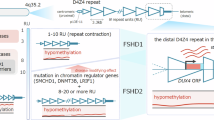
Abstract
In recent years, whole-exome and whole-genome sequencing have been increasingly applied for the genetic diagnosis of muscle diseases. However, standard short-read sequencing often fails to detect pathogenic variants in some inherited muscle diseases, such as Duchenne/Becker muscular dystrophy (DMD/BMD), facioscapulohumeral muscular dystrophy (FSHD), oculopharyngeal muscular dystrophy (OPMD), and oculopharyngodistal myopathy (OPDM). This review outlines the genetic diagnostic approaches for these conditions, with a particular focus on novel analytical approaches for genetic diagnosis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Nishikawa A, Mitsuhashi S, Miyata N, Nishino I. Targeted massively parallel sequencing and histological assessment of skeletal muscles for the molecular diagnosis of inherited muscle disorders. J Med Genet. 2017;54:104–10.
Zhong J, Xie Y, Bhandari V, Chen G, Dang Y, Liao H, et al. Clinical and genetic characteristics of female dystrophinopathy carriers. Mol Med Rep. 2019;19:3035–44.
Okubo M, Minami N, Goto K, Goto Y, Noguchi S, Mitsuhashi S, et al. Genetic diagnosis of Duchenne/Becker muscular dystrophy using next-generation sequencing: validation analysis of DMD mutations. J Hum Genet. 2016;61:483–9.
Okubo M, Noguchi S, Hayashi S, Nakamura H, Komaki H, Matsuo M, et al. Exon skipping induced by nonsense/frameshift mutations in DMD gene results in Becker muscular dystrophy. Hum Genet. 2020;139:247–55.
Okubo M, Noguchi S, Awaya T, Hosokawa M, Tsukui N, Ogawa M, et al. RNA-seq analysis, targeted long-read sequencing and in silico prediction to unravel pathogenic intronic events and complicated splicing abnormalities in dystrophinopathy. Hum Genet. 2023;142:59–71.
Matthews E, Brassington R, Kuntzer T, Jichi F, Manzur AY. Corticosteroids for the treatment of Duchenne muscular dystrophy. Cochrane Database Syst Rev. 2016;2016:CD003725.
Koeks Z, Bladen CL, Salgado D, van Zwet E, Pogoryelova O, McMacken G, et al. Clinical outcomes in duchenne muscular dystrophy: a study of 5345 patients from the TREAT-NMD DMD global database. J Neuromuscul Dis. 2017;4:293–306.
Angelini C, Pegoraro E, Turella E, Intino MT, Pini A, Costa C. Deflazacort in Duchenne dystrophy: study of long-term effect. Muscle Nerve. 1994;17:386–91.
Pramono ZA, Takeshima Y, Alimsardjono H, Ishii A, Takeda S, Matsuo M. Induction of exon skipping of the dystrophin transcript in lymphoblastoid cells by transfecting an antisense oligodeoxynucleotide complementary to an exon recognition sequence. Biochem Biophys Res Commun. 1996;226:445–9.
Komaki H, Nagata T, Saito T, Masuda S, Takeshita E, Sasaki M, et al. Systemic administration of the antisense oligonucleotide NS-065/NCNP-01 for skipping of exon 53 in patients with Duchenne muscular dystrophy. Sci Transl Med. 2018;10:eaan0713.
Clemens PR, Rao VK, Connolly AM, Harper AD, Mah JK, McDonald CM, et al. Safety, tolerability, and efficacy of viltolarsen in boys with Duchenne muscular dystrophy amenable to exon 53 skipping: a phase 2 randomized clinical trial. JAMA Neurol. 2020;77:982–91.
Mendell JR, Goemans N, Lowes LP, Alfano LN, Berry K, Shao J, et al. Longitudinal effect of eteplirsen versus historical control on ambulation in Duchenne muscular dystrophy. Ann Neurol. 2016;79:257–71.
Frank DE, Schnell FJ, Akana C, El-Husayni SH, Desjardins CA, Morgan J, et al. Increased dystrophin production with golodirsen in patients with Duchenne muscular dystrophy. Neurology. 2020;94:e2270–e2282.
Zaidman CM, Proud CM, McDonald CM, Lehman KJ, Goedeker NL, Mason S, et al. Delandistrogene moxeparvovec gene therapy in ambulatory patients (aged ≥4 to <8 years) with Duchenne muscular dystrophy: 1-year interim results from Study SRP-9001-103 (ENDEAVOR). Ann Neurol. 2023;94:955–68.
US Food and Drug Administration. FDA investigating deaths due to acute liver failure following treatment with Sarepta’s AAVrh74 gene therapies. 2025 Jun 24. Available at: https://www.fda.gov/vaccines-blood-biologics/safety-availability-biologics/fda-investigating-deaths-due-acute-liver-failure-following-treatment-sareptas-aavrh74-gene-therapies. Accessed 1 Aug 2025.
US Food and Drug Administration. FDA requests Sarepta Therapeutics suspend distribution of Elevidys and places clinical trials on hold. 2025 Jul 18. Available at: https://www.fda.gov/news-events/press-announcements/fda-requests-sarepta-therapeutics-suspend-distribution-elevidys-and-places-clinical-trials-hold. Accessed 1 Aug 2025.
McDonald CM, Campbell C, Torricelli RE, Finkel RS, Flanigan KM, Goemans N, et al. Safety and efficacy of ataluren in patients with nonsense mutation DMD: a multicentre, randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;390:1489–98.
Birnkrant DJ, Bushby K, Bann CM, Apkon SD, Blackwell A, Colvin MK, et al. Diagnosis and management of Duchenne muscular dystrophy, part 2: respiratory, cardiac, bone health, and orthopaedic management. Lancet Neurol. 2018;17:347–61.
Chemello F, Chai AC, Li H, Rodriguez-Caycedo C, Sanchez-Ortiz E, Bassel-Duby R, et al. Precise correction of Duchenne muscular dystrophy exon deletion mutations by base and prime editing. Sci Adv. 2021;7:eabg4910.
Tabebordbar M, Zhu K, Cheng JK, Chew WL, Widrick JJ, Yan WX, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–11.
Skuk D, Goulet M, Roy B, Chapdelaine P, Bouchard JP, Roy R, et al. Dystrophin expression in muscles of Duchenne muscular dystrophy patients after high-density injections of normal myogenic cells. J Neuropathol Exp Neurol. 2006;65:371–86.
van Geel M, Dickson MC, Beck AF, Bolland DJ, Frants RR, van der Maarel SM, et al. Genomic analysis of human chromosome 10q and 4q telomeres suggests a common origin. Genomics. 2002;79:210–7.
Lemmers RJ, de Kievit P, Sandkuijl LA, Padberg GW, van Ommen GJ, Frants RR, et al. Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat Genet. 2002;32:235–6.
Lemmers RJ, van der Vliet PJ, Klooster R, Sacconi S, Camaño P, Dauwerse JG, et al. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science. 2010;329:1650–3.
Bakker E, Wijmenga C, Vossen RH, Padberg GW, Hewitt J, van der Wielen M, et al. The FSHD-linked locus D4F104S1 (p13E-11) on 4q35 has a homologue on 10qter. Muscle Nerve Suppl 2, S39–S44 (1995).
Deidda G, Cacurri S, Grisanti P, Vigneti E, Piazzo N, Felicetti L. Physical mapping evidence for a duplicated region on chromosome 10qter showing high homology with the facioscapulohumeral muscular dystrophy locus on chromosome 4qter. Eur J Hum Genet. 1995;3:155–67.
Lemmers RJ, Tawil R, Petek LM, Balog J, Block GJ, Santen GW, et al. Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat Genet. 2012;44:1370–4.
van den Boogaard ML, Lemmers RJLF, Balog J, Wohlgemuth M, Auranen M, Mitsuhashi S, et al. Mutations in DNMT3B modify epigenetic repression of the D4Z4 repeat and the penetrance of facioscapulohumeral dystrophy. Am J Hum Genet. 2016;98:1020–9.
Hamanaka K, Šikrová D, Mitsuhashi S, Masuda H, Sekiguchi Y, Sugiyama A, et al. Homozygous nonsense variant in LRIF1 associated with facioscapulohumeral muscular dystrophy. Neurology. 2020;94:e2441–e2447.
van Deutekom JC, Bakker E, Lemmers RJ, van der Wielen MJ, Bik E, Hofker MH, et al. Evidence for subtelomeric exchange of 3.3 kb tandemly repeated units between chromosomes 4q35 and 10q26: implications for genetic counselling and etiology of FSHD1. Hum Mol Genet. 1996;5:1997–2003.
Jones TI, Yan C, Sapp PC, McKenna-Yasek D, Kang PB, Quinn C, et al. Identifying diagnostic DNA methylation profiles for facioscapulohumeral muscular dystrophy in blood and saliva using bisulfite sequencing. Clin Epigenetics. 2014;6:23.
van Overveld PG, Lemmers RJ, Sandkuijl LA, Enthoven L, Winokur ST, Bakels F, et al. Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat Genet. 2003;35:315–7.
de Greef JC, Lemmers RJ, van Engelen BGM, Sacconi S, Venance SL, Frants RR, et al. Common epigenetic changes of D4Z4 in contraction-dependent and contraction-independent FSHD. Hum Mutat. 2009;30:1449–59.
Hamanaka K, Goto K, Arai M, Nagao K, Obuse C, Noguchi S, et al. Clinical, muscle pathological, and genetic features of Japanese facioscapulohumeral muscular dystrophy 2 (FSHD2) patients with SMCHD1 mutations. Neuromuscul Disord. 2016;26:300–8.
Hiramuki Y, Kure Y, Saito Y, Ogawa M, Ishikawa K, Mori-Yoshimura M, et al. Simultaneous measurement of the size and methylation of chromosome 4qA-D4Z4 repeats in facioscapulohumeral muscular dystrophy by long-read sequencing. J Transl Med. 2022;20:517.
Wallace LM, Garwick SE, Mei W, Belayew A, Coppee F, Ladner KJ, et al. DUX4, a candidate gene for facioscapulohumeral muscular dystrophy, causes p53-dependent myopathy in vivo. Ann Neurol. 2011;69:540–52.
Bosnakovski D, Gearhart MD, Toso EA, Recht OO, Cucak A, Jain AK, et al. p53-independent DUX4 pathology in cell and animal models of facioscapulohumeral muscular dystrophy. Dis Model Mech. 2017;10:1211–6.
Himeda CL, Jones TI, Virbasius CM, Zhu LJ, Green MR, Jones PL. Identification of epigenetic regulators of DUX4-fl for targeted therapy of facioscapulohumeral muscular dystrophy. Mol Ther. 2018;26:1797–807.
Ogasawara M, Eura N, Iida A, Kumutpongpanich T, Minami N, Nonaka I, et al. Intranuclear inclusions in muscle biopsy can differentiate oculopharyngodistal myopathy and oculopharyngeal muscular dystrophy. Acta Neuropathol Commun. 2022;10:176.
Brais B, Bouchard JP, Xie YG, Rochefort DL, Chrétien N, Tomé FM, et al. Short GCG expansions in the PABP2 gene cause oculopharyngeal muscular dystrophy. Nat Genet. 1998;18:164–7.
Richard P, Trollet C, Gidaro T, Demay L, Brochier G, Malfatti E, et al. PABPN1 (GCN)11 as a Dominant Allele in Oculopharyngeal Muscular Dystrophy -Consequences in Clinical Diagnosis and Genetic Counselling. J Neuromuscul Dis. 2015;2:175–80.
Richard P, Trollet C, Stojkovic T, de Becdelievre A, Perie S, Pouget J, et al. Correlation between PABPN1 genotype and disease severity in oculopharyngeal muscular dystrophy. Neurology. 2017;88:359–65.
Müller T, Deschauer M, Kolbe-Fehr F, Zierz S. Genetic heterogeneity in 30 German patients with oculopharyngeal muscular dystrophy. J Neurol. 2006;253:892–5.
Abu-Baker A, Kharma N, Perreault J, Grant A, Shekarabi M, Maios C, et al. RNA-based therapy utilizing oculopharyngeal muscular dystrophy transcript knockdown and replacement. Mol Ther Nucleic Acids. 2019;15:12–25.
Robinson DO, Wills AJ, Hammans SR, Read SP, Sillibourne J. Oculopharyngeal muscular dystrophy: a point mutation which mimics the effect of the PABPN1 gene triplet repeat expansion mutation. J Med Genet. 2006;43:e23.
Robinson DO, Hilton-Jones D, Mansfield D, Hildebrand GD, Marks S, Mechan D, et al. Two cases of oculopharyngeal muscular dystrophy (OPMD) with the rare PABPN1 c.35G>C; p.Gly12Ala point mutation. Neuromuscul Disord. 2011;21:809–11.
Ishiura H, Shibata S, Yoshimura J, Suzuki Y, Qu W, Doi K, et al. Noncoding CGG repeat expansions in neuronal intranuclear inclusion disease, oculopharyngodistal myopathy and an overlapping disease. Nat Genet. 2019;51:1222–32.
Deng J, Yu J, Li P, Luan X, Cao L, Zhao J, et al. Expansion of GGC repeat in GIPC1 is associated with oculopharyngodistal myopathy. Am J Hum Genet. 2020;106:793–804.
Sone J, Mitsuhashi S, Fujita A, Mizuguchi T, Hamanaka K, Mori K, et al. Long-read sequencing identifies GGC repeat expansions in NOTCH2NLC associated with neuronal intranuclear inclusion disease. Nat Genet. 2019;51:1215–21.
Yu J, Shan J, Yu M, Di L, Xie Z, Zhang W, et al. The CGG repeat expansion in RILPL1 is associated with oculopharyngodistal myopathy type 4. Am J Hum Genet. 2022;109:533–41.
Eura N, Iida A, Ogasawara M, Hayashi S, Noguchi S, Nishino I. RILPL1-related OPDM is absent in a Japanese cohort. Am J Hum Genet. 2022;109:2088–9.
Liufu T, Zheng Y, Yu J, Yuan Y, Wang Z, Deng J, et al. The polyG diseases: a new disease entity caused by CGG repeat expansions in noncoding regions of multiple genes. Acta Neuropathol Commun. 2022;10:79.
Green KM, Glineburg MR, Kearse MG, Flores BN, Linsalata AE, Fedak SJ, et al. RAN translation at C9orf72-associated repeat expansions is selectively enhanced by the integrated stress response. Nat Commun. 2017;8:2005.
Wright SE, Rodriguez CM, Monroe J, Xing J, Krans A, Flores BN, et al. CGG repeats trigger translational frameshifts that generate aggregation-prone chimeric proteins. Nucleic Acids Res. 2022;50:8674–89.
Acknowledgements
We thank all the members of the Medical Genome Center at the NCNP for their daily efforts in analyzing the genetic data that formed the basis of this review.
Funding
This study was supported partly by Intramural Research Grant (5-6, 5-5) for Neurological and Psychiatric Disorders of NCNP; Health, Labor, and Welfare Sciences Research Grants (JPMH23FC1014); and JPMH24FC1009 Matsumura Group: “Research for the Dissemination of Standardized Medical Care for Muscular Dystrophy”. Thank you for your attention.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Saito, Y., Nishino, I. Disease-specific genetic diagnostic strategies for muscle diseases unresolved by short-read sequencing. J Hum Genet (2025). https://doi.org/10.1038/s10038-025-01391-5
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s10038-025-01391-5