Abstract
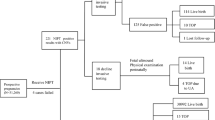
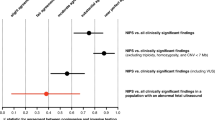
Non-invasive prenatal testing (NIPT) enables the screening of fetal chromosomal abnormalities by analyzing cell-free DNA (cfDNA) in maternal blood. Recent technological advancements have expanded its applications to the detection of copy number variations (CNVs). However, the clinical utility of CNV detection remains unclear. We aimed to investigate the association between fetal CNVs detected by genome-wide NIPT and perinatal outcomes in a large cohort in Japan. This retrospective cohort study included 46,082 patients who underwent NIPT at certified facilities in Japan between January 2015 and September 2021. Genome-wide NIPT was performed using massively parallel sequencing to detect fetal CNVs exceeding 7 Mb. Despite their small size, well-characterized microdeletions, such as 22q11.2 were included. From 46,082 patients with NIPT results, 30,373 cases with known birth outcomes were extracted, and cases with fetal CNV were included in the analysis. Fetal CNVs were detected in 66 patients (0.2%). Adverse outcomes, including miscarriage, growth restriction, and structural abnormalities, were observed in 14 of the 66 cases (21.2%). Pathogenic CNVs were frequently detected even in the 52 cases (78.8%) with favorable outcomes. Genome-wide NIPT may assist in the diagnosis of cases with structural abnormalities when combined with confirmatory testing. Our findings demonstrate that pathogenic CNVs are also detected in a substantial number of structurally normal fetuses with favorable short-term outcomes. This discordance presents a significant challenge for prenatal counseling. The clinical significance of the findings should be clarified through confirmatory testing of CNV cases and the accumulation of data from long-term follow-up studies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Bianchi DW, Chiu RWK. Sequencing of circulating cell-free DNA during pregnancy. N Engl J Med. 2018;379:464–73.
Bianchi DW, Platt LD, Goldberg JD, Abuhamad AZ, Sehnert AJ, Rava RP, et al. Genome-wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstet Gynecol. 2012;119:890–901.
Palomaki GE, Kloza EM, Lambert-Messerlian GM, Haddow JE, Neveux LM, Ehrich M, et al. DNA sequencing of maternal plasma to detect Down syndrome: an international clinical validation study. Genet Med. 2011;13:913–20.
Benn P, Cuckle H, Pergament E. Non-invasive prenatal testing for aneuploidy: current status and future prospects. Ultrasound Obstet Gynecol. 2013;4:15–33.
Chitty LS, Bianchi DW. Noninvasive prenatal testing: the paradigm is shifting rapidly. Prenat Diagn. 2013;33:511–3.
Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350:485–7.
Suzumori N, Sekizawa A, Takeda E, Samura O, Sasaki A, Akaishi R, et al. Classification of factors involved in nonreportable results of noninvasive prenatal testing (NIPT) and prediction of success rate of second NIPT. Prenat Diagn. 2019;39:100–6.
Sago H, Sekizawa A, J.N. Consortium. Nationwide demonstration project of next-generation sequencing of cell-free DNA in maternal plasma in Japan: 1-year experience. Prenat Diagn. 2015;35:331–6.
Suzumori N, Sekizawa A, Tajeda E, Samura O, Sasaki A, Akaishi R, et al. Retrospective details of false-positive and false-negative results in non-invasive prenatal testing for fetal trisomies 21, 18 and 13. Eur J Obstet Gynecol Reprod Biol. 2021;256:75–81.
Takahashi M, Linh LK, Sayed AM, Imoto A, Sato M, Dila KAS, et al. Non-Invasive prenatal testing (NIPT) implementation in Japan: a comparison with the United Kingdom, Germany, Italy, Sweden, and Taiwan. Int J Environ Res Public Health. 2022;19:16404.
Yamada T, Sekizawa A, Fujii Y, Hirose T, Samura O, Suzumori N, et al. Maternal age-specific risk for trisomy 21 based on the clinical performance of NIPT and empirically derived NIPT age-specific positive and negative predictive values in Japan. J Hum Genet. 2018;63:1035–40.
Suzumori N. What are the ethical issues involved in noninvasive prenatal testing in Japan? J Obstet Gynaecol Res. 2022;48:300–5.
Samura O, Sekizawa A, Suzumori N, Sasaki A, Wada S, Hamanoue H, et al. Current status of non-invasive prenatal testing in Japan. J Obstet Gynaecol Res. 2017;43:1245–55.
Samura O. Update on noninvasive prenatal testing: a review based on current worldwide research. J Obstet Gynaecol Res. 2020;46:1246–54.
Connor C, Sato T, Bianchi DW, Fenton K, Somani E, Turriff A, et al. Comparing the introduction and implementation of noninvasive prenatal testing (NIPT) in Japan, the Netherlands, and the United States: an integrative review. Prenat Diagn. 2025;45:1244–64.
American College of Obstetricians and Gynecologists’ Committee on Practice Bulletins—Obstetrics, Committee on Genetics, & Society for Maternal-Fetal Medicine. Screening for fetal chromosomal abnormalities: ACOG Practice Bulletin, Number 226. Obstet Gynecol. 2020;136:e48–69.
Soster E, Tynan J, Gibbons C, Meschino W, Wardrop J, Almasri E, et al. Laboratory performance of genome-wide cfDNA for copy number variants as compared to prenatal microarray. Mol Cytogenet. 2023;16:10.
Pynaker C, Morris F, Hui L, Halliday J. Perinatal outcomes and genomic characteristics of fetal copy number variants: an individual record linkage study of 713 pregnancies. Prenat Diagn. 2023;43:516–26.
Wang W, Lu F, Zhang B, Zhou Q, Chen Y, Yu B. Clinical evaluation of non-invasive prenatal screening for the detection of fetal genome-wide copy number variants. Orphanet J Rare Dis. 2022;17:253.
Kanda Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transpl. 2013;48:452–8.
Yan X, Ding K, Zhang X, Zhang S, Peng H, Zhang Y. Analysis of prenatal diagnosis and pregnancy outcomes for rare autosomal trisomies detected by non-invasive prenatal testing in 33,079 cases. BMC Med Genomics. 2025;18:29.
Raymond Y, Shavi F, Menezes M, Mol BW, McLennan A, da Silva Costa F, et al. Placental, maternal, fetal, and technical origins of false-positive cell-free DNA screening results. Am J Obstet Gynecol. 2024;230:381–9.
Spinella F, Biricik A, Minasi M, Greco E, Fiorentino F. Extent of chromosomal mosaicism influences the clinical outcome of in vitro fertilization treatments. Fertil Steril. 2018;109:77–83.
Eggenhuizen GM, Go A, Koster MPH, Baart EB, Galjaard RJ. Confined placental mosaicism and the association with pregnancy outcome and fetal growth: a review of the literature. Hum Reprod Update. 2021;27:885–903.
Riggs ER, Anderson EF, Cherry AM, Kantarci S, Kearney H, Patel A, et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet Med. 2020;22:245–57.
Izumi K, Krantz ID. Pallister-Killian syndrome. Am J Med Genet C Semin Med Genet. 2014;166c:406–13.
Karaman B, Kayserili H, Ghanbari A, Uyguner ZO, Toksoy G, Altunoglu U, et al. Pallister-Killian syndrome: clinical, cytogenetic and molecular findings in 15 cases. Mol Cytogenet. 2018;11:45.
Wang C, Tang J, Tong K, Huang D, Tu H, Li Q, et al. Expanding the application of non-invasive prenatal testing in the detection of foetal chromosomal copy number variations. BMC Med Genomics. 2021;14:292.
Acknowledgements
We thank Dr. Jun Murotsuki (formerly of the Department of Obstetrics and Gynecology, Miyagi Children’s Hospital) and Dr. Mika Ito (formerly of the Department of Obstetrics and Gynecology, Toyama University Hospital) for their valuable contributions in providing clinical data during the early phase of this study. We also would like to thank the clinical geneticists, counselors, and staff at all institutions that contributed to this study.
Funding
This study was supported by a Grant-in-Aid for Scientific Research (C) from the Japan Society for the Promotion of Science (JSPS KAKENHI), Grant Number 23K08810.
Author information
Authors and Affiliations
Contributions
Yuka Yamashita, Seiji Wada, Haruhiko Sago, Yuki Ito, Osamu Samura, Nobuhiro Suzumori, Hideaki Sawai, Yuko Tamaki, Yukiko Katagiri, Yoshiki Maeda, Hiroko Morisaki, Akira Namba, Yoshimasa Kamei, Junko Yotsumoto, Yuri Hasegawa, Kiyonori Miura, Setsuko Nakayama, and Akihiko Sekizawa conceptualized and designed the study. All co-authors were responsible for the acquisition and management of clinical data. Koshichi Goto performed the CNV analysis and interpreted the genetic findings. Yuka Yamashita and Akihiko sekizawa wrote the first draft of the manuscript. All co-authors critically reviewed and revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yamashita, Y., Shirato, N., Ishii, T. et al. Perinatal outcomes of fetal CNVs detected by genome-wide non-invasive prenatal testing in Japan. J Hum Genet 71, 81–89 (2026). https://doi.org/10.1038/s10038-025-01409-y
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s10038-025-01409-y