Abstract
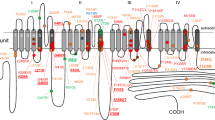
The CACNA1H gene, which encodes the T-type calcium channel Cav3.2, is known to confer susceptibility to childhood absence epilepsy (CAE) and has been implicated in various neurological disorders. However, its pathogenic significance, especially in childhood intractable epilepsies, has not been comprehensively explored. We performed whole-exome sequencing on a 4-year-old boy diagnosed with epilepsy with myoclonic–atonic seizures (EMAtS), and identified two missense variants in CACNA1H. One was a novel variant, p.D949H, inherited from the father, while the other was a known variant, p.R788C, inherited from the mother. Because the latter was previously reported to alter Cav3.2 channel function and contribute to the pathogenesis of CAE and idiopathic generalized epilepsy, we evaluated the former’s functional impact using two-electrode voltage clamp analysis in Xenopus laevis oocytes. While the current–voltage relationship of the D949H mutant channel was not significantly different from that of the wild-type channel, the time constant of recovery from inactivation was significantly prolonged in the D949H variant (671.7 ± 52.0 ms vs. 455.5 ± 28.2 ms), indicating moderately impaired properties of the mutant. Notably, neither the D949H nor the R788C variant was associated with epilepsy in either parent, suggesting that these variants were not sufficient to cause epilepsy on their own, and that the compound heterozygous state of CACNA1H contributed to the EMAtS phenotype in the proband. Our findings highlight the genetic complexity of EMAtS and underscore the importance of accumulated functional impacts of modifier variants in severe epileptic diseases, even when individual variants are not pathogenic.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others

References
Guerrini R, Conti V, Mantegazza M, Balestrini S, Galanopoulou AS, Benfenati F. Developmental and epileptic encephalopathies: from genetic heterogeneity to phenotypic continuum. Physiol Rev. 2023;103:433–513.
Mercimek-Mahmutoglu S, Patel J, Cordeiro D, Hewson S, Callen D, Donner EJ, et al. Diagnostic yield of genetic testing in epileptic encephalopathy in childhood. Epilepsia. 2015;56:707–16.
Helbig KL, Farwell Hagman KD, Shinde DN, Mroske C, Powis Z, Li S, et al. Diagnostic exome sequencing provides a molecular diagnosis for a significant proportion of patients with epilepsy. Genet Med. 2016;18:898–905.
Takata A, Nakashima M, Saitsu H, Mizuguchi T, Mitsuhashi S, Takahashi Y, et al. Comprehensive analysis of coding variants highlights genetic complexity in developmental and epileptic encephalopathy. Nat Commun. 2019;10:2506.
Dulac O. Epileptic encephalopathy. Epilepsia. 2001;42:23–6.
Tang S, Pal DK. Dissecting the genetic basis of myoclonic-astatic epilepsy. Epilepsia. 2012;53:1303–13.
Oguni H. Epilepsy with myoclonic-atonic seizures, also known as Doose syndrome: Modification of the diagnostic criteria. Eur J Paediatr Neurol. 2022;36:37–50.
Carvill GL, McMahon JM, Schneider A, Zemel M, Myers CT, Saykally J, et al. Mutations in the GABA Transporter SLC6A1 Cause Epilepsy with Myoclonic-Atonic Seizures. Am J Hum Genet. 2015;96:808–15.
Chen Y, Lu J, Pan H, Zhang Y, Wu H, Xu K, et al. Association between genetic variation of CACNA1H and childhood absence epilepsy. Ann Neurol. 2003;54:239–43.
Khosravani H, Bladen C, Parker DB, Snutch TP, McRory JE, Zamponi GW. Effects of Cav3.2 channel mutations linked to idiopathic generalized epilepsy. Ann Neurol. 2005;57:745–9.
Scholl UI, Stölting G, Nelson-Williams C, Vichot AA, Choi M, Loring E, et al. Recurrent gain of function mutation in calcium channel CACNA1H causes early-onset hypertension with primary aldosteronism. Elife. 2015;4:e06315.
Vitko I, Chen Y, Arias JM, Shen Y, Wu XR, Perez-Reyes E. Functional characterization and neuronal modeling of the effects of childhood absence epilepsy variants of CACNA1H, a T-type calcium channel. J Neurosci. 2005;25:4844–55.
Heron SE, Khosravani H, Varela D, Bladen C, Williams TC, Newman MR, et al. Extended spectrum of idiopathic generalized epilepsies associated with CACNA1H functional variants. Ann Neurol. 2007;62:560–8.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60.
Cheng J, Novati G, Pan J, Bycroft C, Žemgulytė A, Applebaum T, et al. Accurate proteome-wide missense variant effect prediction with AlphaMissense. Science. 2023;381:eadg7492.
Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, et al. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am J Hum Genet. 2016;99:877–85.
Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47:D886–d94.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419.
Hodge RD, Bakken TE, Miller JA, Smith KA, Barkan ER, Graybuck LT, et al. Conserved cell types with divergent features in human versus mouse cortex. Nature. 2019;573:61–8.
Liman ER, Tytgat J, Hess P. Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron. 1992;9:861–71.
Kise Y, Kasuya G, Okamoto HH, Yamanouchi D, Kobayashi K, Kusakizako T, et al. Structural basis of gating modulation of Kv4 channel complexes. Nature. 2021;599:158–64.
Kanda Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transpl. 2013;48:452–8.
Huang J, Fan X, Jin X, Lyu C, Guo Q, Liu T, et al. Structural basis for human Ca(v)3.2 inhibition by selective antagonists. Cell Res. 2024;34:440–50.
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–9.
Stringer RN, Cmarko L, Zamponi GW, De Waard M, Weiss N. Electrophysiological characterization of a Ca(v)3.2 calcium channel missense variant associated with epilepsy and hearing loss. Mol Brain. 2023;16:68.
Stringer RN, Jurkovicova-Tarabova B, Souza IA, Ibrahim J, Vacik T, Fathalla WM, et al. De novo SCN8A and inherited rare CACNA1H variants associated with severe developmental and epileptic encephalopathy. Mol Brain. 2021;14:126.
Becker F, Reid CA, Hallmann K, Tae HS, Phillips AM, Teodorescu G, et al. Functional variants in HCN4 and CACNA1H may contribute to genetic generalized epilepsy. Epilepsia Open. 2017;2:334–42.
Souza IA, Gandini MA, Zhang FX, Mitchell WG, Matsumoto J, Lerner J, et al. Pathogenic Cav3.2 channel mutation in a child with primary generalized epilepsy. Mol Brain. 2019;12:86.
Weiss N, Zamponi GW. Genetic T-type calcium channelopathies. J Med Genet. 2020;57:1–10.
Polack PO, Guillemain I, Hu E, Deransart C, Depaulis A, Charpier S. Deep layer somatosensory cortical neurons initiate spike-and-wave discharges in a genetic model of absence seizures. J Neurosci. 2007;27:6590–9.
Miyamoto H, Tatsukawa T, Shimohata A, Yamagata T, Suzuki T, Amano K, et al. Impaired cortico-striatal excitatory transmission triggers epilepsy. Nat Commun. 2019;10:1917.
Acknowledgements
We thank the proband and his family for participating in the study.
Funding
This work was supported by JSPS KAKENHI (Grant Numbers. 20K08265 and 23K07254 to A.M. and 21K06786 and 24K02211 to K.N.), the Foundation for Development of the Community, and the Kawano Masanori Memorial Public Interest Incorporated Foundation for the Promotion of Pediatrics (Grant Number 35-5).
Author information
Authors and Affiliations
Contributions
T.M., K.W., M.K., K.M., and H.O. performed clinical data collection and analysis. A.M., S.T., Y.H. and K.M. performed exome sequencing and genetic data interpretation. G.K. and K.N. performed TEVC recordings and analyzed the data. A.M. and T.M. drafted the manuscript. K.N., H.O., and T.M. conceived, designed, and jointly supervised the study. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Matsumoto, A., Kasuya, G., Tumurbaatar, S. et al. Compound heterozygous variants of CACNA1H change channel properties and contribute to intractable epilepsy with myoclonic-atonic seizures. J Hum Genet (2025). https://doi.org/10.1038/s10038-025-01434-x
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s10038-025-01434-x