Abstract
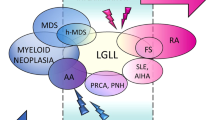
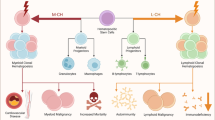
Past studies described occasional patients with myeloid neoplasms (MN) and coexistent large granular lymphocytic leukemia (LGLL) or T-cell clonopathy of unknown significance (TCUS), which may represent expansion of myeloid clonal hematopoiesis (CH) as triggers or targets of clonal cytotoxic T cell reactions. We retrospectively analyzed 349 LGLL/TCUS patients, 672 MN patients, and 1443 CH individuals to establish the incidence, genetic landscape, and clinical phenotypes of CH in LGLL. We identified 8% of cases overlapping with MN, while CH was found in an additional 19% of cases (CH + /LGLL) of which TET2 (23%) and DNMT3A (14%) were the most common. In MN cohort, 3% of cases showed coexistent LGLL. The incidence of CH in LGLL was exceedingly higher than age-matched CH controls (P < 0.0001). By multivariate analysis, the presence of CH in LGLL (P = 0.026) was an independent risk factor for cytopenia in addition to older age (P = 0.003), splenomegaly (P = 0.015) and STAT3/5B mutations (P = 0.001). CH + /LGLL cases also showed a higher progression rate to MN than CH-/LGLL (10% vs. 2% at 5 years; P = 0.02). A close relationship between CH and LGLL suggests that cytopenia in LGLL may be not only related to LGLL but be also secondary to coexisting clonal cytopenia of unclear significance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data to reproduce our study are presented in the manuscript and supplemental material. Additional information can be requested, if needed, by e-mailing the corresponding author.
References
Loughran TP Jr., Kadin ME, Starkebaum G, Abkowitz JL, Clark EA, Disteche C, et al. Leukemia of large granular lymphocytes: association with clonal chromosomal abnormalities and autoimmune neutropenia, thrombocytopenia, and hemolytic anemia. Ann Intern Med. 1985;102:169–75.
Loughran TP Jr, Starkebaum G. Clinical features in large granular lymphocytic leukemia. Blood. 1987;69:1786.
Lamy T, Moignet A, Loughran TP Jr. LGL leukemia: from pathogenesis to treatment. Blood. 2017;129:1082–94.
Sanikommu SR, Clemente MJ, Chomczynski P, Afable MG 2nd, Jerez A, et al. Clinical features and treatment outcomes in large granular lymphocytic leukemia (LGLL). Leuk Lymphoma. 2018;59:416–22.
Viny AD, Lichtin A, Pohlman B, Loughran T, Maciejewski J. Chronic B-cell dyscrasias are an important clinical feature of T-LGL leukemia. Leuk Lymphoma. 2008;49:932–8.
Pawarode A, Baer M. T/B and not T/B: high frequency of B-cell dyscrasias in T-LGL leukemia. Leuk Lymphoma. 2008;49:845–6.
Sidiqi MH, Aljama MA, Viswanatha DS, Dingli D. T-cell large granular lymphocytic leukemia and plasma cell disorders. Haematologica. 2019;104:e108–10.
Sokol L, Loughran TP Jr. Large granular lymphocyte leukemia. Oncologist. 2006;11:263–73.
Komrokji RS, Ali NA, Sallman D, Padron E, Lancet J, Sokol L, et al. Characterization of myelodysplastic syndromes (MDS) with T-cell large granular lymphocyte proliferations (LGL). Leukemia. 2020;34:3097–9.
Zhang X, Sokol L, Bennett JM, Moscinski LC, List A, Zhang L. T-cell large granular lymphocyte proliferation in myelodysplastic syndromes: Clinicopathological features and prognostic significance. Leuk Res. 2016;43:18–23.
Saunthararajah Y, Molldrem JL, Rivera M, Williams A, Stetler-Stevenson M, Sorbara L, et al. Coincident myelodysplastic syndrome and T-cell large granular lymphocytic disease: clinical and pathophysiological features. Br J Haematol. 2001;112:195–200.
Epling-Burnette PK, Painter JS, Rollison DE, Ku E, Vendron D, Widen R, et al. Prevalence and clinical association of clonal T-cell expansions in Myelodysplastic Syndrome. Leukemia. 2007;21:659–67.
Bravo-Perez C, Gurnari C. A tower of babel of acronyms? The shadowlands of MGUS/MBL/CHIP/TCUS. Semin Hematol. 2024;61:43–50.
Swerdlow SH CE, Harris NL, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, vol. 2. IARC Press, 2017.
Poullot E, Zambello R, Leblanc F, Bareau B, De March E, Roussel M, et al. Chronic natural killer lymphoproliferative disorders: characteristics of an international cohort of 70 patients. Ann Oncol. 2014;25:2030–5.
Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–98.
Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20:1472–8.
Uddin MDM, Nguyen NQH, Yu B, Brody JA, Pampana A, Nakao T, et al. Clonal hematopoiesis of indeterminate potential, DNA methylation, and risk for coronary artery disease. Nat Commun. 2022;13:5350.
Buscarlet M, Provost S, Zada YF, Barhdadi A, Bourgoin V, Lépine G, et al. DNMT3A and TET2 dominate clonal hematopoiesis and demonstrate benign phenotypes and different genetic predispositions. Blood. 2017;130:753–62.
Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303.
Gurnari C, Wahida A, Pagliuca S, Durmaz A, Zawit M, Haferlach T, et al. A study of Telomerase Reverse Transcriptase rare variants in myeloid neoplasia. Hematol Oncol. 2022;40:812–7.
Adema V, Palomo L, Walter W, Mallo M, Hutter S, La Framboise T, et al. Pathophysiologic and clinical implications of molecular profiles resultant from deletion 5q. EBioMedicine. 2022;80:104059.
Kongkiatkamon S, Pagliuca S, Adema V, Nagata Y, Kerr CM, Walter W, et al. Molecular characterization of the histone acetyltransferase CREBBP/EP300 genes in myeloid neoplasia. Leukemia. 2022;36:1185–8.
Cheon H, Xing JC, Moosic KB, Ung J, Chan VW, Chung DS, et al. Genomic landscape of TCRαβ and TCRγδ T-large granular lymphocyte leukemia. Blood. 2022;139:3058–72.
Olson TL, Cheon H, Xing JC, Olson KC, Paila U, Hamele CE, et al. Frequent somatic TET2 mutations in chronic NK-LGL leukemia with distinct patterns of cytopenias. Blood. 2021;138:662–73.
Baer C, Kimura S, Rana MS, Kleist AB, Flerlage T, Feith DJ, et al. CCL22 mutations drive natural killer cell lymphoproliferative disease by deregulating microenvironmental crosstalk. Nat Genet. 2022;54:637–48.
Niroula A, Sekar A, Murakami MA, Trinder M, Agrawal M, Wong WJ, et al. Distinction of lymphoid and myeloid clonal hematopoiesis. Nat Med. 2021;27:1921–7.
Genovese G, Kahler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–87.
Khoury JD, Solary E, Abla O, Akkari Y, Alaggio R, Apperley JF, et al. The 5th edition of the World Health Organization classification of haematolymphoid tumours: myeloid and histiocytic/dendritic neoplasms. Leukemia. 2022;36:1703–19.
Nagata Y, Makishima H, Kerr CM, Przychodzen BP, Aly M, Goyal A, et al. Invariant patterns of clonal succession determine specific clinical features of myelodysplastic syndromes. Nat Commun. 2019;10:5386.
Bernstein N, Spencer Chapman M, Nyamondo K, Chen Z, Williams N, Mitchell E, et al. Analysis of somatic mutations in whole blood from 200,618 individuals identifies pervasive positive selection and novel drivers of clonal hematopoiesis. Nat Genet. 2024;56:1147–55.
Beauchamp EM, Leventhal M, Bernard E, Hoppe ER, Todisco G, Creignou M, et al. ZBTB33 is mutated in clonal hematopoiesis and myelodysplastic syndromes and impacts RNA splicing. Blood Cancer Discov. 2021;2:500–17.
Slavin TP, Teh JB, Weitzel JN, Peng K, Wong FL, Qin H, et al. Association between clonal hematopoiesis and late nonrelapse mortality after autologous hematopoietic cell transplantation. Biol Blood Marrow Transplant. 2019;25:2517–21.
Kewan T, Durmaz A, Bahaj W, Gurnari C, Terkawi L, Awada H, et al. Molecular patterns identify distinct subclasses of myeloid neoplasia. Nat Commun. 2023;14:3136.
Koskela HL, Eldfors S, Ellonen P, van Adrichem AJ, Kuusanmäki H, Andersson EI, et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med. 2012;366:1905–13.
Barilà G, Grassi A, Cheon H, Teramo A, Calabretto G, Chahal J, et al. Tγδ LGLL identifies a subset with more symptomatic disease: analysis of an international cohort of 137 patients. Blood. 2023;141:1036–46.
Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24.
Huerga Encabo H, Aramburu IV, Garcia-Albornoz M, Piganeau M, Wood H, Song A, et al. Loss of TET2 in human hematopoietic stem cells alters the development and function of neutrophils. Cell Stem Cell. 2023;30:781–99.e789.
Kahles A, Lehmann KV, Toussaint NC, Hüser M, Stark SG, Sachsenberg T, et al. Comprehensive analysis of alternative splicing across tumors from 8705 patients. Cancer Cell. 2018;34:211–24.e216.
Biernacki MA, Lok J, Black RG, Foster KA, Cummings C, Woodward KB, et al. Discovery of U2AF1 neoantigens in myeloid neoplasms. J Immunother Cancer. 2023;11:e007490.
Acknowledgements
This work was supported by the R35HL135795 (to J.P.M.); the Edward P. Evans Foundation (to C.G.); Case Comprehensive Cancer Center and VeloSano Bike to Cure Award (to C.G. & A.D.) and AA&MDSIF (to C.B-P., V.V., J.P.M); VeloSano 9 Pilot Award (to V.V.). N.K. has a postdoctoral fellowship from Astellas Foundation for Research on Metabolic Disorders and the Uehara Memorial Foundation. C.B-P. has a postdoctoral fellowship from Instituto de Salud Carlos III (JR22/00041).
Author information
Authors and Affiliations
Contributions
N.K. supervised the study, collected, analyzed, interpreted clinical and molecular data and wrote the manuscript; C.G., C.B-P. L.G. and V.V. collected, analyzed, interpreted clinical and molecular data and edited the manuscript. Y.K. and A.D. analyzed molecular data, S.P., N.W., A.A., D.D. and F.U. collected clinical and molecular data. H.E.C. and A.S interpreted clinical and molecular data and edited the manuscript. J.P.M. provided invaluable help with the manuscript preparation, generated and conceived the study design, designed figures and tables, and wrote the manuscript. All authors participated in the critical review of the final paper and submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Ethics approval and consent to participate
This research included human subjects in accordance with the Declaration of Helsinki. Informed consent was obtained from patients after approval by the Internal review board (Irb 5024 and 15–1278).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kawashima, N., Gurnari, C., Bravo-Perez, C. et al. Clonal hematopoiesis in large granular lymphocytic leukemia. Leukemia 39, 451–459 (2025). https://doi.org/10.1038/s41375-024-02460-y
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41375-024-02460-y
This article is cited by
-
ASXL1 mutation-related clonal hematopoiesis and age-related diseases: clinical evidence and molecular insights
International Journal of Hematology (2025)